5MTZ
 
 | | Crystal structure of a long form RNase Z from yeast | | 分子名称: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | 著者 | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | 登録日 | 2017-01-11 | | 公開日 | 2017-06-21 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
1JJ0
 
 | |
2MKO
 
 | | G-triplex structure and formation propensity | | 分子名称: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | 著者 | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | 登録日 | 2014-02-11 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
5ULD
 
 | |
2MDZ
 
 | | NMR structure of the Paracoccus denitrificans Z-subunit determined in the presence of ADP | | 分子名称: | Uncharacterized protein | | 著者 | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2013-09-20 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
6HLK
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | 分子名称: | Redirecting phage packaging protein C (RppC) | | 著者 | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | 登録日 | 2018-09-11 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
7NK5
 
 | |
7NM1
 
 | |
2M1I
 
 | |
7NLE
 
 | |
6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | 著者 | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | 登録日 | 2017-10-21 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
7NLA
 
 | |
6AQT
 
 | |
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | 分子名称: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | 著者 | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | 登録日 | 2001-09-25 | | 公開日 | 2001-11-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
6AQW
 
 | |
7NM9
 
 | |
7KER
 
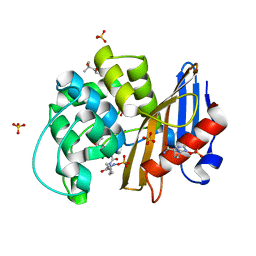 | | avibactam-CDD-1 45 minute complex | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-10-12 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-05-26 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
2M35
 
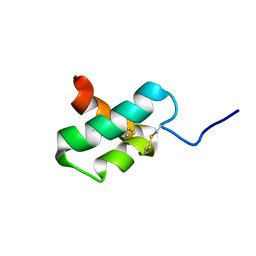 | | NMR study of k-Ssm1a | | 分子名称: | k-Ssm1a | | 著者 | King, G.F, Undheim, E.A, Mobli, M, Yang, S, Rong, M, Lai, R. | | 登録日 | 2013-01-09 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR study of k-Ssm1a
To be Published
|
|
6AR6
 
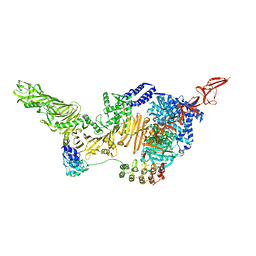 | |
7NK3
 
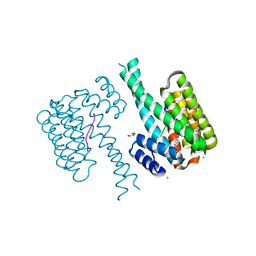 | |
7NJ9
 
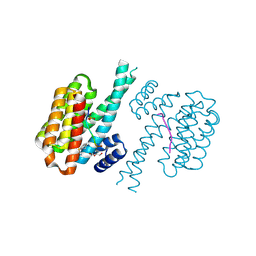 | |
7NJB
 
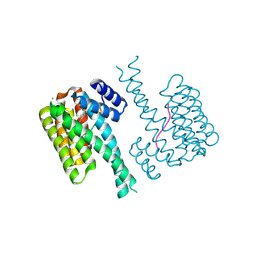 | |
7NIF
 
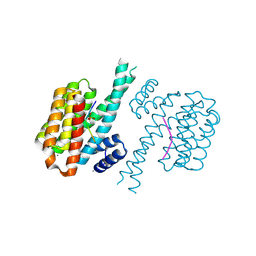 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-011 | | 分子名称: | 1-(4-methylphenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-02-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7KIS
 
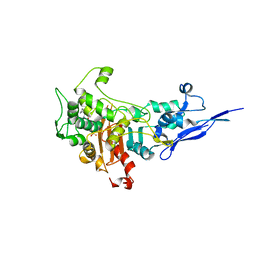 | | Crystal structure of Pseudomonas aeruginosa PBP2 in complex with WCK 5153 | | 分子名称: | (2S,5R)-1-formyl-N'-[(3R)-pyrrolidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Peptidoglycan D,D-transpeptidase MrdA | | 著者 | Rajavel, M, van den Akker, F. | | 登録日 | 2020-10-24 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.869 Å) | | 主引用文献 | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7KNH
 
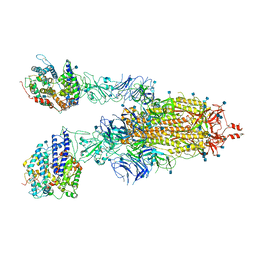 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
