6LLZ
 
 | |
6U5F
 
 | | CryoEM Structure of Pyocin R2 - precontracted - collar | | 分子名称: | Collar PA0615, Sheath PA0622, Tube PA0623 | | 著者 | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | 登録日 | 2019-08-27 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | 著者 | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | 登録日 | 2021-10-02 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95000446 Å) | | 主引用文献 | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
1WFX
 
 | | Crystal Structure of APE0204 from Aeropyrum pernix | | 分子名称: | CHLORIDE ION, Probable RNA 2'-phosphotransferase, ZINC ION | | 著者 | Kato-Murayama, M, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-27 | | 公開日 | 2004-11-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the RNA 2'-Phosphotransferase from Aeropyrum pernix K1
J.Mol.Biol., 348, 2005
|
|
7NIG
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1008 | | 分子名称: | 14-3-3 protein sigma, 2-bromanyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-02-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6OQR
 
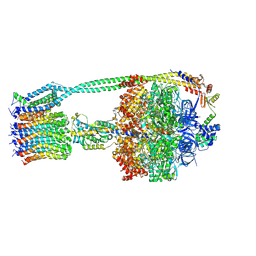 | | E. coli ATP Synthase ADP State 1a | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Stewart, A.G, Walshe, J.L, Sobti, M. | | 登録日 | 2019-04-29 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7NKU
 
 | | diazaborine bound Drg1(AFG2) | | 分子名称: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ATPase family gene 2 protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Prattes, M, Bergler, H, Haselbach, D. | | 登録日 | 2021-02-19 | | 公開日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine.
Nat Commun, 12, 2021
|
|
6LU8
 
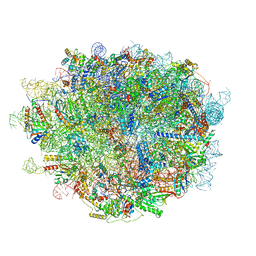 | | Cryo-EM structure of a human pre-60S ribosomal subunit - state A | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | 登録日 | 2020-01-26 | | 公開日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
7NJA
 
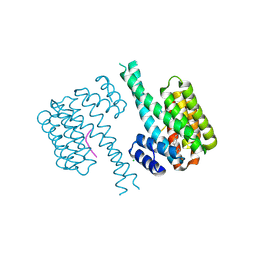 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1006 | | 分子名称: | 14-3-3 protein sigma, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-02-16 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
4OVT
 
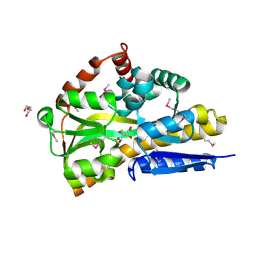 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTERIUM ANTHROPI (Oant_3902), TARGET EFI-510153, WITH BOUND L-FUCONATE | | 分子名称: | 6-deoxy-L-galactonic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-14 | | 公開日 | 2014-01-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
8OOY
 
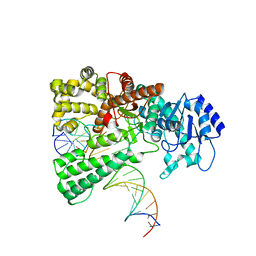 | | Pol I bound to extended and displaced DNA section - open conformation | | 分子名称: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | 著者 | Botto, M, Borsellini, A, Lamers, M.H. | | 登録日 | 2023-04-06 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1WHV
 
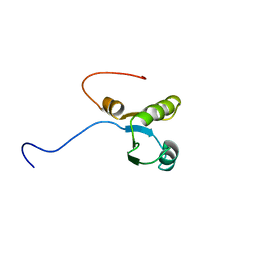 | | Solution structure of the RNA binding domain from hypothetical protein BAB23382 | | 分子名称: | poly(A)-specific ribonuclease | | 著者 | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RNA binding domain from hypothetical protein BAB23382
To be Published
|
|
1X1V
 
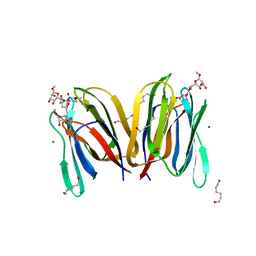 | | Structure Of Banana Lectin- Methyl-Alpha-Mannose Complex | | 分子名称: | HEXANE-1,6-DIOL, ZINC ION, lectin, ... | | 著者 | Singh, D.D, Saikrishnan, K, Kumar, P, Surolia, A, Sekar, K, Vijayan, M. | | 登録日 | 2005-04-14 | | 公開日 | 2005-11-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Unusual sugar specificity of banana lectin from Musa paradisiaca and its probable evolutionary origin. Crystallographic and modelling studies
Glycobiology, 15, 2005
|
|
6M0S
 
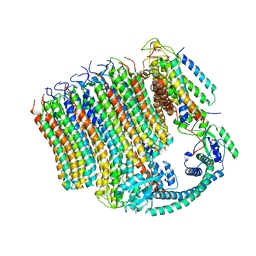 | | 3.6A Yeast Vo state3 prime | | 分子名称: | Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | 著者 | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, SIngharoy, A, Chiu, W. | | 登録日 | 2020-02-22 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
1J10
 
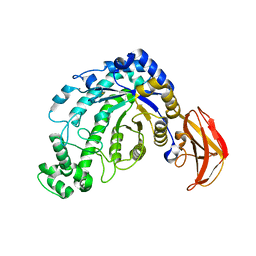 | | beta-amylase from Bacillus cereus var. mycoides in complex with GGX | | 分子名称: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-xylopyranose, ... | | 著者 | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | 登録日 | 2002-11-25 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
6I2G
 
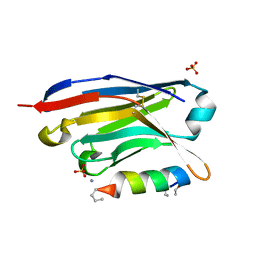 | |
6M3Q
 
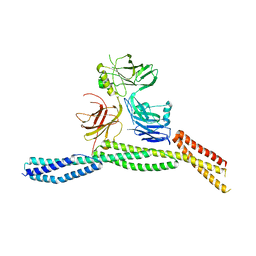 | | Crystal structure of AnkB/beta4-spectrin complex | | 分子名称: | Ankyrin-2, Spectrin beta chain | | 著者 | Li, J, Chen, K, Zhu, R, Zhang, M. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.436 Å) | | 主引用文献 | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
6O8Q
 
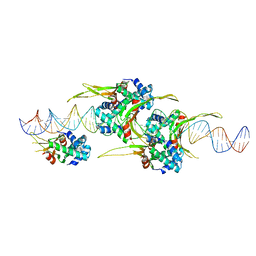 | | HUaa 19bp SYM DNA pH 4.5 | | 分子名称: | DNA (57-MER), DNA-binding protein HU-alpha | | 著者 | Remesh, S.G, Hammel, M. | | 登録日 | 2019-03-11 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.216 Å) | | 主引用文献 | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
1J21
 
 | |
6I93
 
 | |
8B9X
 
 | | Chimeric protein of human UFM1 E3 ligase, UFL1, and DDRGK1 | | 分子名称: | CHLORIDE ION, DDRGK domain-containing protein 1,E3 UFM1-protein ligase 1 | | 著者 | Wiener, R, Isupov, M, banerjee, S. | | 登録日 | 2022-10-10 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.066 Å) | | 主引用文献 | Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.
Embo Rep., 24, 2023
|
|
6M4Q
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | 登録日 | 2020-03-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
7PHP
 
 | | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | | 分子名称: | Anti-Fab nanobody, Multidrug resistance protein NorM, NabFab HC, ... | | 著者 | Bloch, J.S, Mukherjee, S, Kowal, J, Niederer, M, Pardon, E, Steyaert, J, Kossiakoff, A.A, Locher, K.P. | | 登録日 | 2021-08-18 | | 公開日 | 2021-09-01 | | 最終更新日 | 2021-12-01 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1X4C
 
 | | Solution structure of RRM domain in splicing factor 2 | | 分子名称: | Splicing factor, arginine/serine-rich 1 | | 著者 | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-14 | | 公開日 | 2005-11-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of RRM domain in splicing factor 2
To be Published
|
|
6RFX
 
 | | Crystal structure of Eis2 from Mycobacterium abscessus | | 分子名称: | ACETATE ION, CITRIC ACID, Eis2, ... | | 著者 | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | 登録日 | 2019-04-16 | | 公開日 | 2019-07-10 | | 最終更新日 | 2019-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
