5V17
 
 | |
5V7Y
 
 | |
5G4H
 
 | | 1.50 A resolution catechol (1,2-dihydroxybenzene) inhibited Sporosarcina pasteurii urease | | 分子名称: | 1,2-ETHANEDIOL, CATECHOL, HYDROXIDE ION, ... | | 著者 | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | 登録日 | 2016-05-13 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Inactivation of Urease by Catechol: Kinetics and Structure.
J.Inorg.Biochem., 166, 2016
|
|
6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | 分子名称: | Portal protein | | 著者 | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
7KEO
 
 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | 分子名称: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | 著者 | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | 登録日 | 2020-10-11 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
1Y84
 
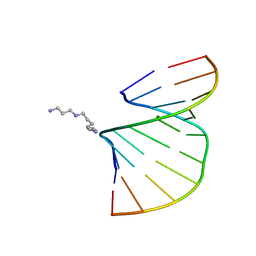 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-(imidazolyl)ethyl] Thymidine (T*) | | 分子名称: | 5'-D(*GP*CP*GP*TP*AP*(EIT)P*AP*CP*GP*C)-3', MAGNESIUM ION, SPERMINE | | 著者 | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | 登録日 | 2004-12-10 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
8DTN
 
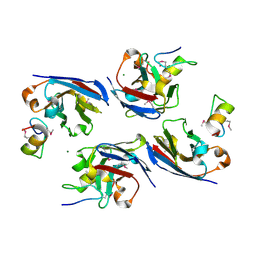 | | The complex of nanobody 6101 with BCL11A ZF6 | | 分子名称: | B-cell lymphoma/leukemia 11A, MAGNESIUM ION, Nanobody 6101, ... | | 著者 | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | 登録日 | 2022-07-26 | | 公開日 | 2023-02-01 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1YAO
 
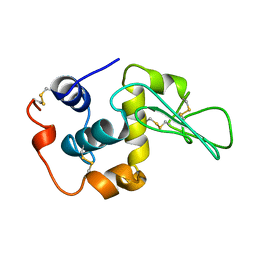 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | 登録日 | 1995-09-29 | | 公開日 | 1996-04-03 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
6CJ0
 
 | |
1LQJ
 
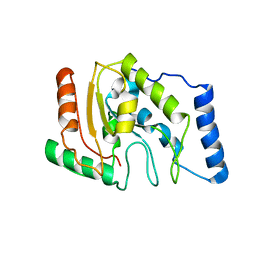 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | 分子名称: | URACIL-DNA GLYCOSYLASE | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8DTU
 
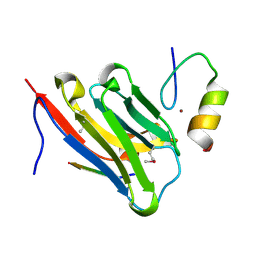 | | The complex of nanobody 5344N74D with BCL11A ZF6. | | 分子名称: | B-cell lymphoma/leukemia 11A, Nanobody 5344N74D, ZINC ION | | 著者 | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | 登録日 | 2022-07-26 | | 公開日 | 2023-02-01 | | 実験手法 | X-RAY DIFFRACTION (2.447 Å) | | 主引用文献 | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1Y43
 
 | | crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | 分子名称: | Aspergillopepsin II heavy chain, Aspergillopepsin II light chain, SULFATE ION | | 著者 | Sasaki, H, Nakagawa, A, Iwata, S, Muramatsu, T, Suganuma, M, Sawano, Y, Kojima, M, Kubota, K, Takahashi, K. | | 登録日 | 2004-11-30 | | 公開日 | 2005-12-13 | | 最終更新日 | 2013-02-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger
Proc.Jpn.Acad.,Ser.B, 80, 2004
|
|
3OK8
 
 | | I-BAR OF PinkBAR | | 分子名称: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 2, GLYCEROL | | 著者 | Boczkowska, M, Rebowski, G, Saarikangas, J, Lappalainen, P, Dominguez, R. | | 登録日 | 2010-08-24 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6RUM
 
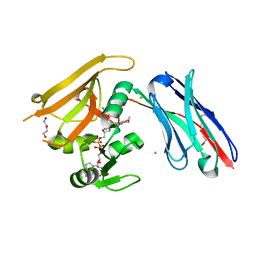 | | Crystal structure of GFP-LAMA-G97 - a GFP enhancer nanobody with cpDHFR insertion and TMP and NADPH | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GFP-LAMA-G97 a GFP enhancer nanobody with cpDHFR insertion, ... | | 著者 | Farrants, H, Tarnawski, M, Mueller, T.G, Otsuka, S, Hiblot, J, Koch, B, Kueblbeck, M, Kraeusslich, H.-G, Ellenberg, J, Johnsson, K. | | 登録日 | 2019-05-28 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Chemogenetic Control of Nanobodies.
Nat.Methods, 17, 2020
|
|
5BRU
 
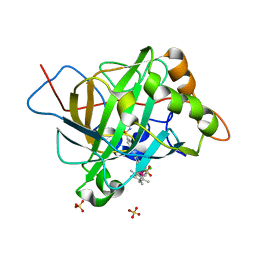 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | 分子名称: | Carbonic anhydrase 2, SULFATE ION, ZINC ION, ... | | 著者 | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | 登録日 | 2015-06-01 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
1LS2
 
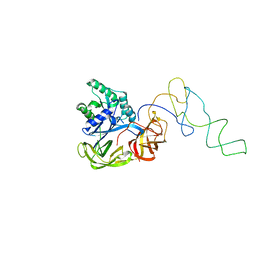 | | Fitting of EF-Tu and tRNA in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | 分子名称: | Elongation Factor Tu, Phenylalanine transfer RNA | | 著者 | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | 登録日 | 2002-05-16 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16.799999 Å) | | 主引用文献 | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
1Y0H
 
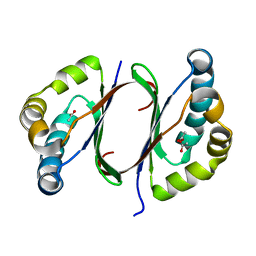 | | Structure of Rv0793 from Mycobacterium tuberculosis | | 分子名称: | ACETATE ION, hypothetical protein Rv0793 | | 著者 | Lemieux, M.J, Ference, C, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2004-11-15 | | 公開日 | 2004-12-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
3OUQ
 
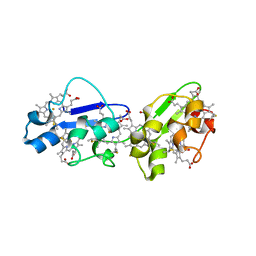 | |
3OV0
 
 | |
6FR2
 
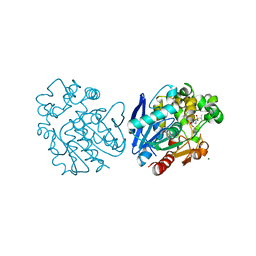 | | Soluble epoxide hydrolase in complex with LK864 | | 分子名称: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | 著者 | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | 登録日 | 2018-02-15 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.262 Å) | | 主引用文献 | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
1CYC
 
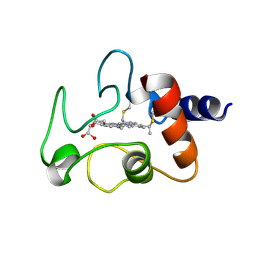 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | 分子名称: | FERROCYTOCHROME C, HEME C | | 著者 | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | 登録日 | 1976-08-01 | | 公開日 | 1976-10-06 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
1RPO
 
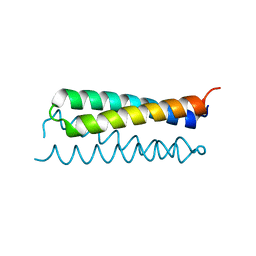 | |
6TH8
 
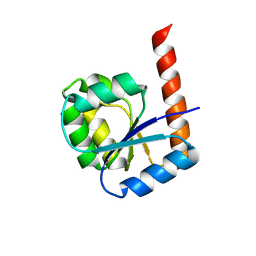 | |
1RSV
 
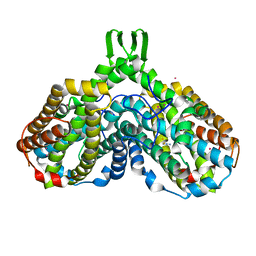 | | azide complex of the diferrous E238A mutant R2 subunit of ribonucleotide reductase | | 分子名称: | AZIDE ION, FE (III) ION, MERCURY (II) ION, ... | | 著者 | Assarsson, M, Andersson, M.E, Hogbom, M, Persson, B.O, Sahlin, M, Barra, A.L, Sjoberg, B.M, Nordlund, P, Graslund, A. | | 登録日 | 2003-12-10 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Restoring proper radical generation by azide binding to the iron site of the E238A mutant R2 protein of ribonucleotide reductase from Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
6TI6
 
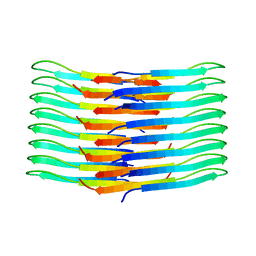 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | 登録日 | 2019-11-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
