7V06
 
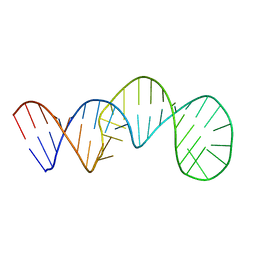 | |
2KYD
 
 | | RDC and RCSA refinement of an A-form RNA: Improvements in Major Groove Width | | Descriptor: | RNA (5'-R(*CP*UP*AP*GP*UP*UP*AP*GP*CP*UP*AP*AP*CP*UP*AP*G)-3') | | Authors: | Tolbert, B.S, Summers, M.F, Miyazaki, Y, Barton, S, Kinde, B, Stark, P, Singh, R, Bax, A, Case, D. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement.
J.Biomol.Nmr, 47, 2010
|
|
2IRO
 
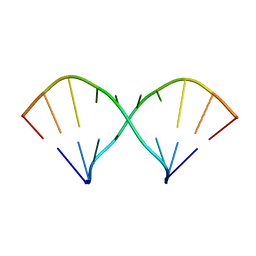 | | The NMR Structures of (rGCUGAGGCU)2 and (rGCGGAUGCU)2 | | Descriptor: | 5'-R(P*GP*CP*GP*GP*AP*UP*GP*CP*U)-3' | | Authors: | Tolbert, B.S. | | Deposit date: | 2006-10-16 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of (rGCUGAGGCU)(2) and (rGCGGAUGCU)(2): Probing the Structural Features That Shape the Thermodynamic Stability of GA Pairs(,).
Biochemistry, 46, 2007
|
|
2IRN
 
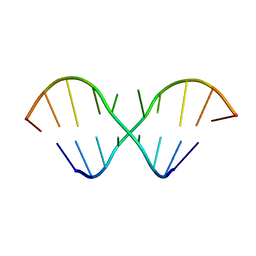 | | The NMR Structures of (rGCUGAGGCU)2 and (rGCGGAUGCU)2 | | Descriptor: | 5'-R(P*GP*CP*UP*GP*AP*GP*GP*CP*U)-3' | | Authors: | Tolbert, B.S. | | Deposit date: | 2006-10-15 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of (rGCUGAGGCU)(2) and (rGCGGAUGCU)(2): Probing the Structural Features That Shape the Thermodynamic Stability of GA Pairs(,).
Biochemistry, 46, 2007
|
|
2N4L
 
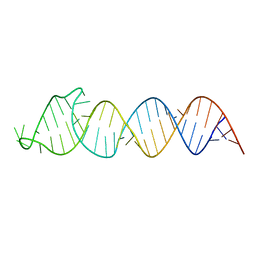 | | Solution Structure of the HIV-1 Intron Splicing Silencer and its Interactions with the UP1 Domain of hnRNP A1 | | Descriptor: | RNA (53-MER) | | Authors: | Tolbert, B.S, Jain, N, Morgan, C.E, Rife, B.D, Salemi, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Intron Splicing Silencer and Its Interactions with the UP1 Domain of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) A1.
J.Biol.Chem., 291, 2016
|
|
6XB7
 
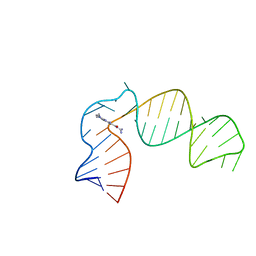 | | IRES-targeting Small Molecule Inhibits Enterovirus 71 Replication via Allosteric Stabilization of a Ternary Complex | | Descriptor: | 3-amino-N-(diaminomethylidene)-5-(dimethylamino)-6-(phenylethynyl)pyrazine-2-carboxamide, RNA (41-MER) | | Authors: | Tolbert, B.S, Davila, J, Sugarman, A.L, Chiu, L.Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex.
Nat Commun, 11, 2020
|
|
5V16
 
 | |
5V17
 
 | |
