4OPK
 
 | | Bh-RNaseH:2'-SMe-DNA complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*AP*(USM)P*TP*CP*GP*CP*G)-3', GLYCEROL, Ribonuclease H | | 著者 | Pallan, P.S, Egli, M. | | 登録日 | 2014-02-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Generating Crystallographic Models of DNA Dodecamers from Structures of RNase H:DNA Complexes.
Methods Mol.Biol., 1320
|
|
4OQ7
 
 | |
6CUC
 
 | |
4OQI
 
 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S/G238S mutations | | 分子名称: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase | | 著者 | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | 登録日 | 2014-02-09 | | 公開日 | 2015-05-20 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
3KZ3
 
 | |
3KZC
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase | | 分子名称: | N-acetylornithine carbamoyltransferase, SULFATE ION | | 著者 | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2009-12-08 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
J.Biol.Chem., 280, 2005
|
|
3KZM
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate | | 分子名称: | GLYCEROL, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | 著者 | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2009-12-08 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
6CCX
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | 分子名称: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
8VYE
 
 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYG
 
 | | SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
4OV0
 
 | | Structure of Bacteriorhdopsin Transferred from Amphipol A8-35 to a Lipidic Mesophase | | 分子名称: | Bacteriorhodopsin, RETINAL | | 著者 | Polovinkin, V, Gushchin, I, Sintsov, M, Round, E, Balandin, T, Chervakov, P, Schevchenko, V, Utrobin, P, Popov, A, Borshchevskiy, V, Mishin, A, Kuklin, A, Willbold, D, Popot, J.L, Gordeliy, V. | | 登録日 | 2014-02-19 | | 公開日 | 2014-10-01 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase.
J.Membr.Biol., 247, 2014
|
|
4O6K
 
 | | The crystal structure of zebrafish IL-22 | | 分子名称: | Interleukin 22 | | 著者 | Siupka, P, Hamming, O.J, Fretaud, M, Luftalla, G, Levraud, J.P, Hartmann, R. | | 登録日 | 2013-12-21 | | 公開日 | 2014-09-10 | | 最終更新日 | 2018-05-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of zebrafish IL-22 reveals an evolutionary, conserved structure highly similar to that of human IL-22.
Genes Immun., 15, 2014
|
|
3KZO
 
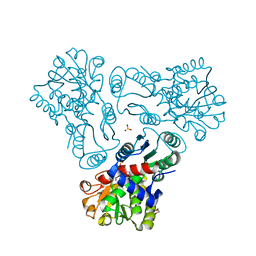 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate and N-acetyl-L-norvaline | | 分子名称: | GLYCEROL, N-ACETYL-L-NORVALINE, N-acetylornithine carbamoyltransferase, ... | | 著者 | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2009-12-08 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | 分子名称: | Putative isochorismatase, SULFATE ION | | 著者 | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-11-06 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
8VYF
 
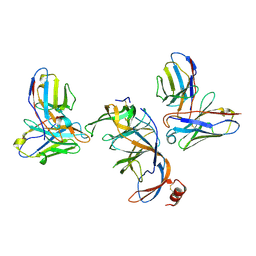 | | SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
4OBG
 
 | |
3L2A
 
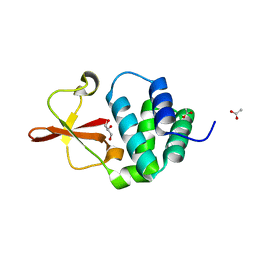 | | Crystal structure of Reston Ebola VP35 interferon inhibitory domain | | 分子名称: | ACETIC ACID, GLYCEROL, Polymerase cofactor VP35 | | 著者 | Leung, D.W, Farahbakhsh, M, Borek, D.M, Prins, K.C, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural and Functional Characterization of Reston Ebola Virus VP35 Interferon Inhibitory Domain.
J.Mol.Biol., 399, 2010
|
|
4OCB
 
 | |
6D8C
 
 | | Cryo-EM structure of FLNaABD E254K bound to phalloidin-stabilized F-actin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Iwamoto, D.V, Huehn, A.R, Simon, B, Huet-Calderwood, C, Baldassarre, M, Sindelar, C.V, Calderwood, D.A. | | 登録日 | 2018-04-26 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural basis of the filamin A actin-binding domain interaction with F-actin.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3KQ4
 
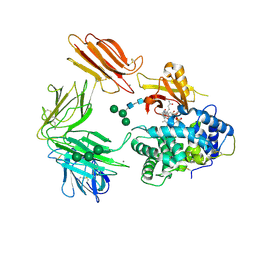 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | 登録日 | 2009-11-17 | | 公開日 | 2010-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
6DBL
 
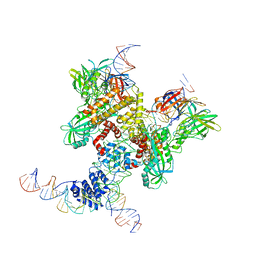 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.001 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DA1
 
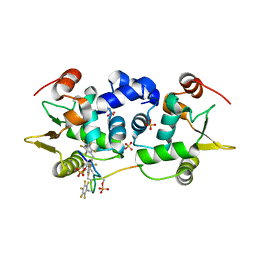 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-01 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.000127 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | 分子名称: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3L1P
 
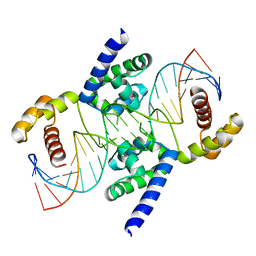 | | POU protein:DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP*TP*CP*AP*AP*AP*TP*GP*TP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP*CP*AP*AP*AP*TP*GP*GP*A)-3'), POU domain, ... | | 著者 | Vahokoski, J, Groves, M.R, Pogenberg, V, Wilmanns, M. | | 登録日 | 2009-12-14 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A unique Oct4 interface is crucial for reprogramming to pluripotency
Nat.Cell Biol., 15, 2013
|
|
