1KDI
 
 | | REDUCED FORM OF PLASTOCYANIN FROM DRYOPTERIS CRASSIRHIZOMA | | 分子名称: | COPPER (II) ION, PLASTOCYANIN | | 著者 | Inoue, T, Gotowda, M, Hamada, K, Kohzuma, T, Yoshizaki, F, Sugimura, Y, Kai, Y. | | 登録日 | 1998-05-08 | | 公開日 | 1999-05-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure and unusual pH dependence of plastocyanin from the fern Dryopteris crassirhizoma. The protonation of an active site histidine is hindered by pi-pi interactions.
J.Biol.Chem., 274, 1999
|
|
3D1G
 
 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | 著者 | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | 登録日 | 2008-05-05 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4J5D
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | 分子名称: | 1-(4-aminobenzyl)-3-{2-[(2R)-2-(2-bromophenyl)pyrrolidin-1-yl]-2-oxoethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | 著者 | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | 登録日 | 2013-02-08 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1KQF
 
 | | FORMATE DEHYDROGENASE N FROM E. COLI | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARDIOLIPIN, FORMATE DEHYDROGENASE, ... | | 著者 | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | 登録日 | 2002-01-05 | | 公開日 | 2002-03-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
4G76
 
 | |
4JQ9
 
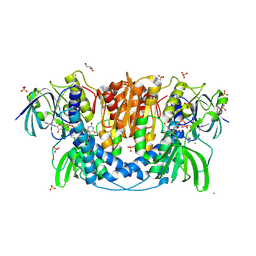 | | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex | | 分子名称: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Tietzel, M, Neumann, P, Meyer, D, Ficner, R, Tittmann, K. | | 登録日 | 2013-03-20 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex
TO BE PUBLISHED
|
|
1KEU
 
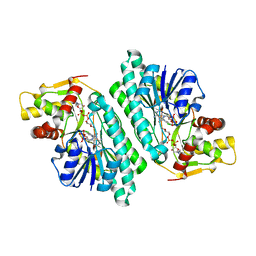 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with dTDP-D-glucose bound | | 分子名称: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-D-glucose 4,6-dehydratase | | 著者 | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | 登録日 | 2001-11-17 | | 公開日 | 2002-01-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6R9T
 
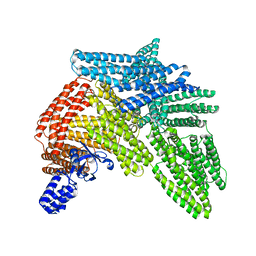 | | Cryo-EM structure of autoinhibited human talin-1 | | 分子名称: | Talin-1 | | 著者 | Dedden, D, Schumacher, S, Zacharias, M, Biertumpfel, C, Mizuno, N. | | 登録日 | 2019-04-04 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | The Architecture of Talin1 Reveals an Autoinhibition Mechanism.
Cell, 179, 2019
|
|
4J61
 
 | |
4ZQM
 
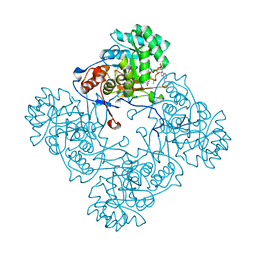 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with XMP and NAD | | 分子名称: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
3CSK
 
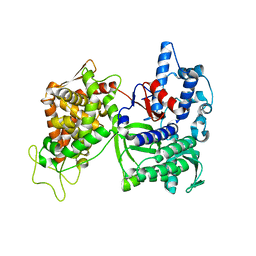 | | Structure of DPP III from Saccharomyces cerevisiae | | 分子名称: | MAGNESIUM ION, Probable dipeptidyl-peptidase 3, ZINC ION | | 著者 | Baral, P.K, Jajcanin, N, Deller, S, Macheroux, P, Abramic, M, Gruber, K. | | 登録日 | 2008-04-10 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding.
J.Biol.Chem., 283, 2008
|
|
4J66
 
 | |
2CB1
 
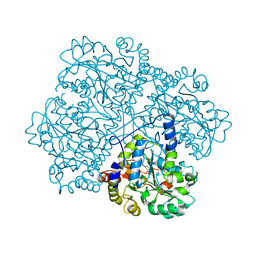 | | Crystal Structure of O-actetyl Homoserine Sulfhydrylase From Thermus Thermophilus HB8,OAH2. | | 分子名称: | O-ACETYL HOMOSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Imagawa, T, Utsunomiya, H, Tsuge, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S. | | 登録日 | 2005-12-28 | | 公開日 | 2007-01-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structure of O-Acetyl Homoserine Sulfhydrylase
To be Published
|
|
6JJU
 
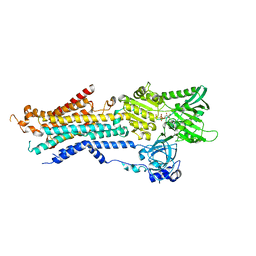 | | Structure of Ca2+ ATPase | | 分子名称: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | 登録日 | 2019-02-27 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
6RBP
 
 | |
1KFB
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH Indole Glycerol Phosphate | | 分子名称: | INDOLE-3-GLYCEROL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | 著者 | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | 登録日 | 2001-11-20 | | 公開日 | 2003-01-07 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | On the Role of alphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1KSM
 
 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | 分子名称: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | 著者 | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | 登録日 | 2002-01-14 | | 公開日 | 2002-01-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
6RBY
 
 | |
6JKC
 
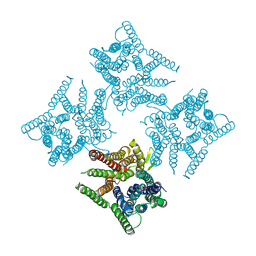 | | Crystal structure of tetrameric PepTSo2 in P4212 space group | | 分子名称: | Proton:oligopeptide symporter POT family | | 著者 | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | 登録日 | 2019-02-28 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1KFN
 
 | |
6G8M
 
 | | Yeast 20S proteasome in complex with Cystargolide B Derivative 1 | | 分子名称: | (2~{S},3~{R})-4-[[(2~{S})-3-methyl-1-[[(2~{S})-3-methyl-1-oxidanylidene-1-phenylmethoxy-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]amino]-3-oxidanyl-4-oxidanylidene-2-propan-2-yl-butanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Groll, M, Tello-Aburto, R. | | 登録日 | 2018-04-09 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
3OK4
 
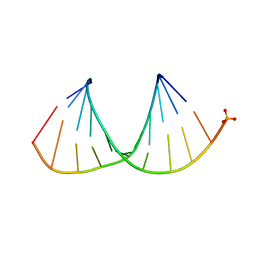 | |
4HD7
 
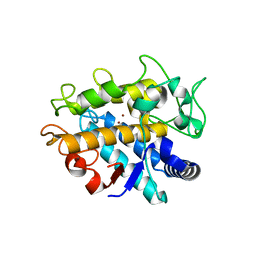 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218G mutant soaked in CuSO4 | | 分子名称: | COPPER (II) ION, Tyrosinase | | 著者 | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | 登録日 | 2012-10-02 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-20 | | 公開日 | 2008-07-15 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
3U4R
 
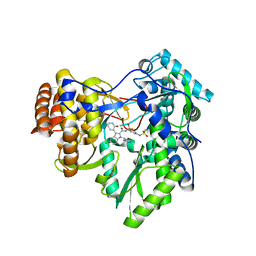 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | 分子名称: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | 著者 | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | 登録日 | 2011-10-10 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
