7JMP
 
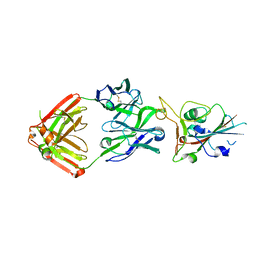 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-39 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-39 heavy chain, COVA2-39 light chain, ... | | 著者 | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | 登録日 | 2020-08-02 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.712 Å) | | 主引用文献 | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
6STK
 
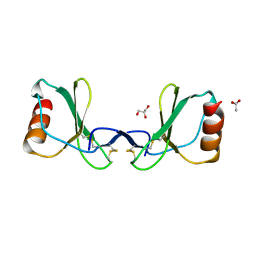 | |
6SVH
 
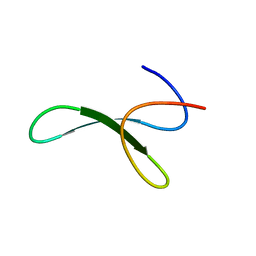 | | Protein allostery of the WW domain at atomic resolution: FFpSPR bound structure | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7EZJ
 
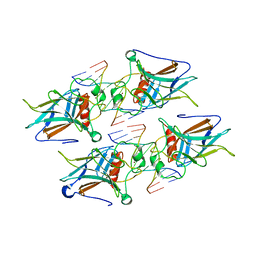 | | Crystal structure of p73 DNA binding domain complex bound with 1 bp and 2 bp spacer DNA response elements. | | 分子名称: | 12-mer DNA, Tumor protein p73, ZINC ION | | 著者 | Koley, T, Roy Chowdhury, S, Kumar, M, Kaur, P, Singh, T.P, Viadiu, H, Ethayathulla, A.S. | | 登録日 | 2021-06-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Deciphering the mechanism of p73 recognition of p53 response elements using the crystal structure of p73-DNA complexes and computational studies.
Int.J.Biol.Macromol., 206, 2022
|
|
7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | 分子名称: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-08-03 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
6SWH
 
 | | Crystal structure of the ternary complex between the type 1 pilus proteins FimC, FimI and FimA from E. coli | | 分子名称: | 1,2-ETHANEDIOL, Chaperone protein FimC, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Giese, C, Puorger, C, Ignatov, O, Weber, M, Scharer, M.A, Capitani, G, Glockshuber, R. | | 登録日 | 2019-09-20 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
7JRO
 
 | | Plant Mitochondrial complex IV from Vigna radiata | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CARDIOLIPIN, ... | | 著者 | Maldonado, M, Letts, J.A. | | 登録日 | 2020-08-12 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
7EZC
 
 | | Adenosine A2a receptor mutant-I92N | | 分子名称: | 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,Soluble cytochrome b562 | | 著者 | Cui, M, Zhou, Q, Yao, D, Zhao, S, Song, G. | | 登録日 | 2021-06-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal structure of a constitutive active mutant of adenosine A 2A receptor.
Iucrj, 9, 2022
|
|
1RER
 
 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | 登録日 | 2003-11-07 | | 公開日 | 2004-01-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
7F1U
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-methionine intermediates | | 分子名称: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, L-methionine gamma-lyase, METHIONINE | | 著者 | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | 登録日 | 2021-06-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
6SYD
 
 | |
7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | 分子名称: | L-methionine gamma-lyase | | 著者 | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | 登録日 | 2021-06-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
1RR2
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to 2-ketobutyric acid | | 分子名称: | 2-KETOBUTYRIC ACID, COBALT (II) ION, transcarboxylase 5S subunit | | 著者 | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | 登録日 | 2003-12-08 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
7F1V
 
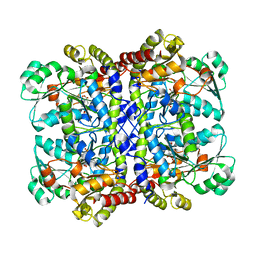 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-homocysteine intermediates | | 分子名称: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, L-methionine gamma-lyase | | 著者 | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | 登録日 | 2021-06-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
1RJ4
 
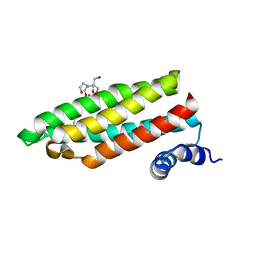 | | Structure of a Cell Wall Invertase Inhibitor from Tobacco in Complex with Cd2+ | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, invertase inhibitor | | 著者 | Hothorn, M, D'Angelo, I, Marquez, J.A, Greiner, S, Scheffzek, K. | | 登録日 | 2003-11-18 | | 公開日 | 2004-02-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension
J.Mol.Biol., 335, 2004
|
|
6T0W
 
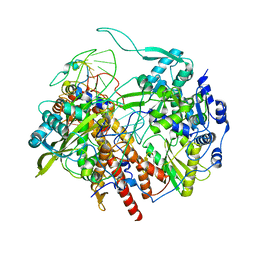 | | Human Influenza B polymerase recycling complex | | 分子名称: | Polymerase PB2, Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit, ... | | 著者 | Wandzik, J.M, Kouba, T, Karuppasamy, M, Cusack, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
7KB2
 
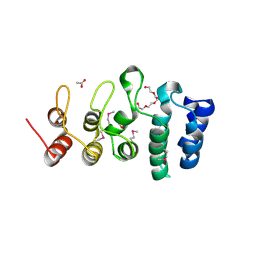 | | Putative ankyrin repeat domain-containing protein from Enterobacter cloacae | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, ANK_REP_REGION domain-containing protein, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-10-01 | | 公開日 | 2020-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Putative ankyrin repeat domain-containing protein from
Enterobacter cloacae
To Be Published
|
|
7K1B
 
 | |
7K39
 
 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Gui, M, Gao, J, Xiang, Y. | | 登録日 | 2020-09-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7JLV
 
 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Disease resistance protein Roq1, MAGNESIUM ION | | 著者 | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | 登録日 | 2020-07-30 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7JMO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-04 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-04 heavy chain, COVA2-04 light chain, ... | | 著者 | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | 登録日 | 2020-08-02 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.359 Å) | | 主引用文献 | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7FEV
 
 | | Crystal structure of Old Yellow Enzyme6 (OYE6) | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN binding | | 著者 | Singh, Y, Sharma, R, Mishra, M, Verma, P.K, Saxena, A.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Crystal structure of ArOYE6 reveals a novel C-terminal helical extension and mechanistic insights into the distinct class III OYEs from pathogenic fungi.
Febs J., 289, 2022
|
|
5OA3
 
 | | Human 40S-eIF2D-re-initiation complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Weisser, M, Schaefer, T, Leibundgut, M, Boehringer, D, Aylett, C.H.S, Ban, N. | | 登録日 | 2017-06-20 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural and Functional Insights into Human Re-initiation Complexes.
Mol. Cell, 67, 2017
|
|
7EXK
 
 | | An AA9 LPMO of Ceriporiopsis subvermispora | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nguyen, H, Kondo, K, Nagata, T, Katahira, M, Mikami, B. | | 登録日 | 2021-05-27 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Functional and Structural Characterizations of Lytic Polysaccharide Monooxygenase, Which Cooperates Synergistically with Cellulases, from Ceriporiopsis subvermispora.
Acs Sustain Chem Eng, 10, 2022
|
|
1YUH
 
 | | FAB FRAGMENT | | 分子名称: | 4-HYDROXY-3-NITROPHENYLACETYL-EPSILON-AMINOCAPROIC ACID, 88C6/12 FAB (HEAVY CHAIN), 88C6/12 FAB (LIGHT CHAIN) | | 著者 | Yuhasz, S.C, Amzel, L.M, Parry, C, Strand, M. | | 登録日 | 1996-01-30 | | 公開日 | 1996-07-11 | | 最終更新日 | 2018-03-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural analysis of affinity maturation: the three-dimensional structures of complexes of an anti-nitrophenol antibody.
Mol.Immunol., 32, 1995
|
|
