3ZKT
 
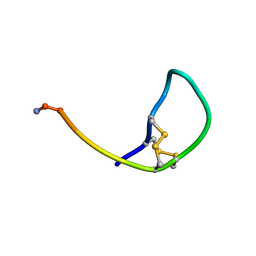 | | SOLUTION STRUCTURE OF THE SOMATOSTATIN SST3 RECEPTOR ANTAGONIST TAU- CONOTOXIN CnVA | | 分子名称: | TAU-CNVA | | 著者 | Petrel, C, Hocking, H.G, Reynaud, M, Favreau, P, Paolini-Bertrand, M, Peigneur, S, Upert, G, Tytgat, J, Gilles, N, Hartley, O, Boelens, R, Stocklin, R, Servent, D. | | 登録日 | 2013-01-24 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification, Structural and Pharmacological Characterization of Tau-Cnva, a Conopeptide that Selectively Interacts with Somatostatin Sst3 Receptor.
Biochem.Pharmacol, 85, 2013
|
|
5EQ9
 
 | |
6W1R
 
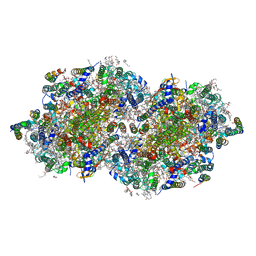 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5EMW
 
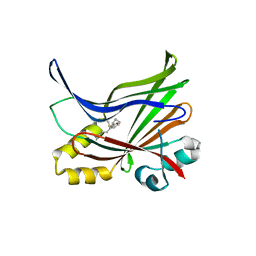 | | Crystal structure of the palmitoylated human TEAD3 transcription factor | | 分子名称: | CALCIUM ION, Transcriptional enhancer factor TEF-5 | | 著者 | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | 登録日 | 2015-11-06 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | 分子名称: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | 登録日 | 2017-07-11 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
6VYI
 
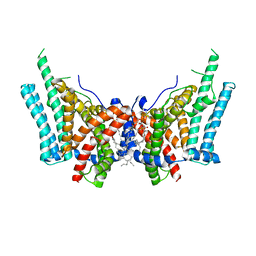 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 | | 分子名称: | Diacylglycerol O-acyltransferase 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese, V.R. | | 登録日 | 2020-02-26 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
6VYT
 
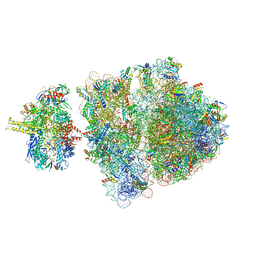 | | Escherichia coli transcription-translation complex A2 (TTC-A2) containing a 15 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Ebright, R.H, Su, M. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5ESA
 
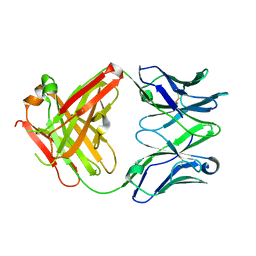 | |
7SCM
 
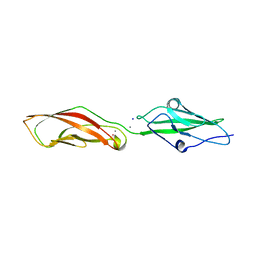 | |
6W69
 
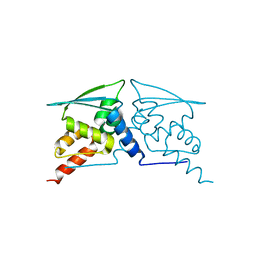 | | The structure of F64, S172A Keap1-BTB domain | | 分子名称: | Kelch-like ECH-associated protein 1 | | 著者 | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | 登録日 | 2020-03-16 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
3ZDT
 
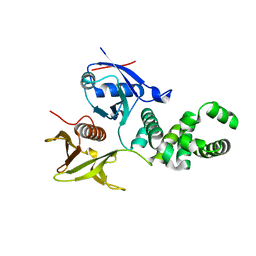 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | 分子名称: | FOCAL ADHESION KINASE 1 | | 著者 | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | 登録日 | 2012-11-30 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5O3J
 
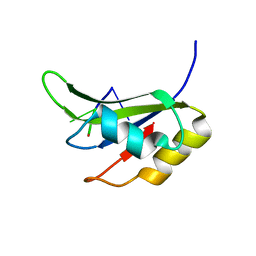 | | Crystal structure of TIA-1 RRM2 in complex with RNA | | 分子名称: | Nucleolysin TIA-1 isoform p40, RNA (5'-R(P*UP*UP*C)-3') | | 著者 | Sonntag, M, Jagtap, P.K.A, Hennig, J, Sattler, M. | | 登録日 | 2017-05-24 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5EOE
 
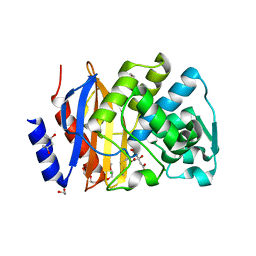 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 (orthorhombic form) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | 登録日 | 2015-11-10 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
3ZBR
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230S, crystallized with NADP | | 分子名称: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | 登録日 | 2012-11-13 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
6VZW
 
 | | TTLL6 bound to the initiation analog | | 分子名称: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, T.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5ESL
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) T322I mutant complexed with itraconazole | | 分子名称: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | 登録日 | 2015-11-16 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of lanosterol 14-alpha demethylase mutants.
To Be Published
|
|
5EP9
 
 | | Crystal structure of the non-heme alpha ketoglutarate dependent epimerase SnoN from nogalamycin biosynthesis | | 分子名称: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | 著者 | Selvaraj, B, Lindqvist, Y, Niiranen, L, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | 登録日 | 2015-11-11 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EQA
 
 | |
7S3T
 
 | | NzeB Diketopiperazine Dimerase Mutant: Q68I-G87A-A89G-I90V | | 分子名称: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Harris, N.R, Shende, V.V, Sanders, J.N, Newmister, S.A, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | 登録日 | 2021-09-08 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 2023
|
|
6W34
 
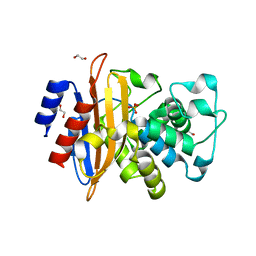 | | Crystal Structure of Class A Beta-lactamase from Bacillus cereus | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-08 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structure of Class A Beta-lactamase from
Bacillus cereus
To Be Published
|
|
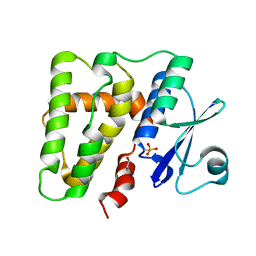
5ELA
 
 | |
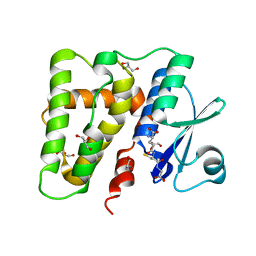
5ELG
 
 | |
5ELK
 
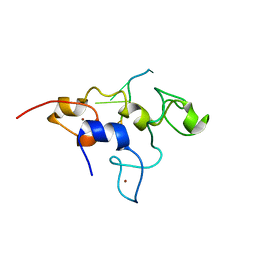 | | Crystal structure of mouse Unkempt zinc fingers 4-6 (ZnF4-6), bound to RNA | | 分子名称: | RING finger protein unkempt homolog, RNA, ZINC ION | | 著者 | Teplova, M, Murn, J, Zarnack, K, Shi, Y, Patel, D.J. | | 登録日 | 2015-11-04 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4JM9
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-amino-1-methylpyridinium | | 分子名称: | 1-METHYL-1,6-DIHYDROPYRIDIN-3-AMINE, Cytochrome c peroxidase, IODIDE ION, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
