5S43
 
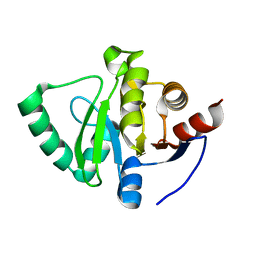 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00024661 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2H
 
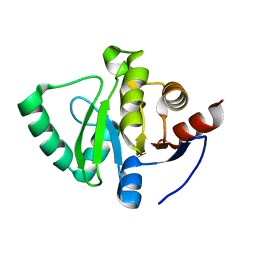 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2856434920 | | Descriptor: | Non-structural protein 3, ethyl (1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)acetate | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.068 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1A5H
 
 | |
5S2X
 
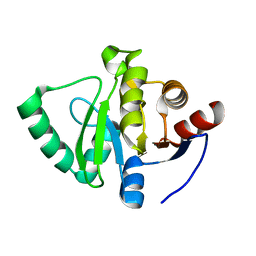 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1139246057 | | Descriptor: | (3R)-N-methyl-1-(pyridazin-3-yl)piperidin-3-amine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S1C
 
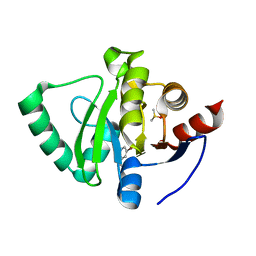 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z3034471507 | | Descriptor: | 1-(5-bromopyridin-3-yl)methanamine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S1O
 
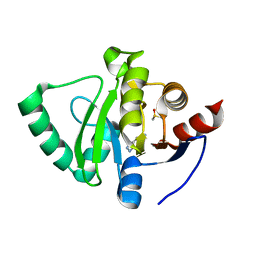 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with STL414928 | | Descriptor: | 2H-pyrazolo[3,4-b]pyridin-5-amine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S24
 
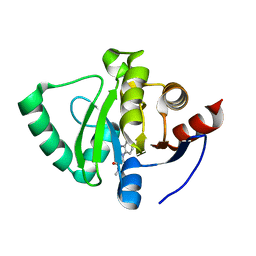 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-697611 | | Descriptor: | 2-(1H-benzimidazol-1-yl)-N-methylacetamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6RFY
 
 | | Crystal structure of Eis2 form Mycobacterium abscessus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Eis2, SULFATE ION | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
5S2N
 
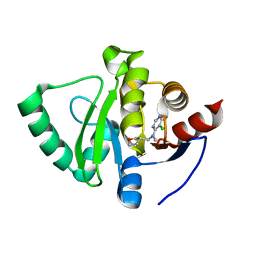 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1787627869 | | Descriptor: | 5-chloranyl-~{N}-methyl-~{N}-[[(3~{S})-oxolan-3-yl]methyl]pyrimidin-4-amine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.133 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S32
 
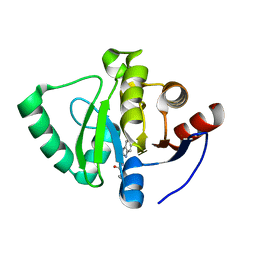 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z26781943 | | Descriptor: | N-[(1H-benzimidazol-2-yl)methyl]-2-methylpropanamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2C
 
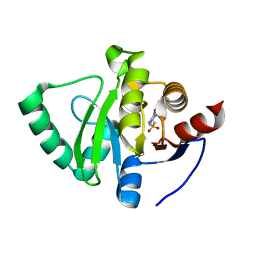 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z45612755 | | Descriptor: | N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)methanesulfonamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.092 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2O
 
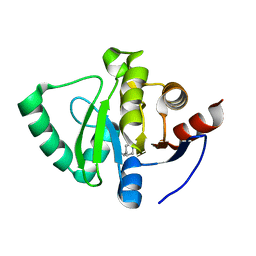 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z645232558 | | Descriptor: | Non-structural protein 3, [1-(pyrimidin-2-yl)piperidin-4-yl]methanol | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.091 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6UN9
 
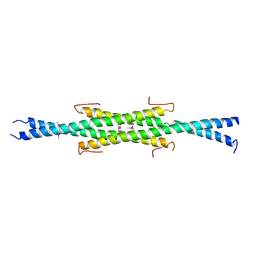 | | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Seetharaman, J, Kolev, M, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2019-10-11 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25
To Be Published
|
|
5S35
 
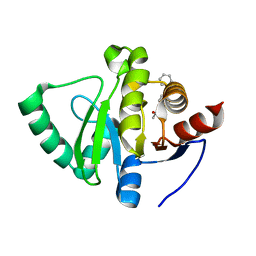 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z68404778 | | Descriptor: | Non-structural protein 3, ~{N}-(4-phenylazanylphenyl)ethanamide | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1HXV
 
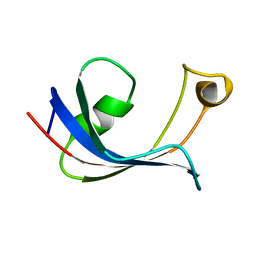 | | PPIASE DOMAIN OF THE MYCOPLASMA GENITALIUM TRIGGER FACTOR | | Descriptor: | TRIGGER FACTOR | | Authors: | Vogtherr, M, Parac, T.N, Maurer, M, Pahl, A, Fiebig, K. | | Deposit date: | 2001-01-17 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and dynamics of the peptidyl-prolyl cis-trans isomerase domain of the trigger factor from Mycoplasma genitalium compared to FK506-binding protein.
J.Mol.Biol., 318, 2002
|
|
3QNN
 
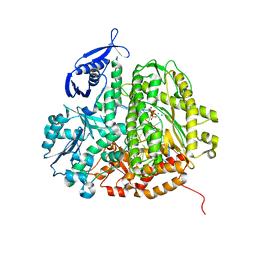 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dGT Opposite 3tCo | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA Primer, ... | | Authors: | Xia, S, Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Using a Fluorescent Cytosine Analogue tC(o) To Probe the Effect of the Y567 to Ala Substitution on the Preinsertion Steps of dNMP Incorporation by RB69 DNA Polymerase.
Biochemistry, 51, 2012
|
|
4ATI
 
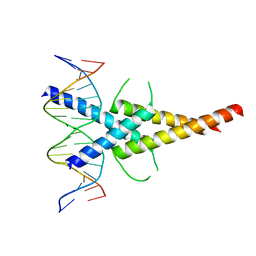 | | MITF:M-box complex | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*CP*AP*TP*GP*TP*GP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*CP*AP*CP*AP*TP*GP*AP*CP*CP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | Authors: | Pogenberg, V, Deineko, V, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
6UO1
 
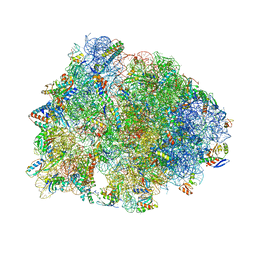 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA (containing pseudouridine at the first position of the codon) and deacylated A-, P-, and E-site tRNAs at 2.95A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Dobosz-Bartoszek, M, Polikanov, Y.S. | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Pseudouridinylation of mRNA coding sequences alters translation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2KGP
 
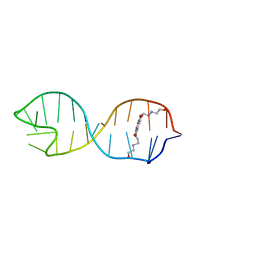 | | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by Novantrone (Mitoxantrone) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, RNA (25-MER) | | Authors: | Zheng, S, Chen, Y, Donahue, C.P, Wolfe, M.S, Varani, G. | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone).
Chem.Biol., 16, 2009
|
|
2BHE
 
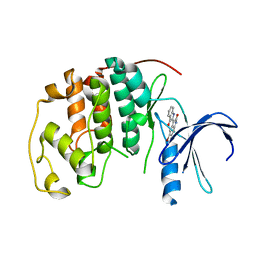 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 5-BROMO-INDIRUBINE | | Descriptor: | (2Z)-5'-BROMO-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE AMMONIATE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-10 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
6ZGZ
 
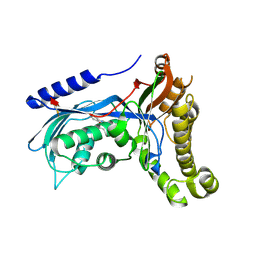 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
2C1L
 
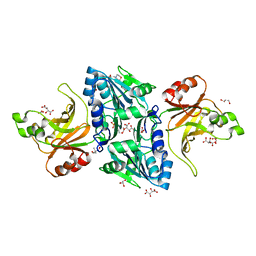 | | Structure of the BfiI restriction endonuclease | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | Authors: | Grazulis, S, Manakova, E, Roessle, M, Bochtler, M, Tamulaitiene, G, Huber, R, Siksnys, V. | | Deposit date: | 2005-09-15 | | Release date: | 2005-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Metal-Independent Restriction Enzyme Bfii Reveals Fusion of a Specific DNA-Binding Domain with a Nonspecific Nuclease.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5OJF
 
 | | Crystal Structure of KLC2-TPR domain (fragment [A1-B6] | | Descriptor: | Kinesin light chain 2 | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Andreani, J, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
1HJI
 
 | | BACTERIOPHAGE HK022 NUN-PROTEIN-NUTBOXB-RNA COMPLEX | | Descriptor: | NUN-PROTEIN, RNA (5-R(P*GP*CP*CP*CP*UP*GP*AP*AP*AP*AP*AP*GP*GP*GP*C)-3) | | Authors: | Faber, C, Schaerpf, M, Becker, T, Sticht, H, Roesch, P. | | Deposit date: | 2001-01-15 | | Release date: | 2002-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Coliphage Hk022 Nun Protein-Lambda-Phage Boxb RNA Complex. Implications for the Mechanism of Transcription Termination
J.Biol.Chem., 276, 2001
|
|
6UCT
 
 | | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Klinke, S, Monti, D, Sycz, G, Cerutti, M.L, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2019-09-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant)
To Be Published
|
|
