8G88
 
 | | Human Oct4 bound to nucleosome with human nMatn1 sequence | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
6NW0
 
 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
6T3C
 
 | | Crystal structure of PI3Kgamma in complex with DNA-PK inhibitor AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6Q84
 
 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
8G8G
 
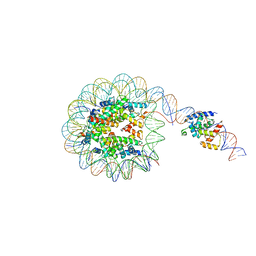 | | Interaction of H3 tail in LIN28B nucleosome with Oct4 | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
6NBC
 
 | |
6Q93
 
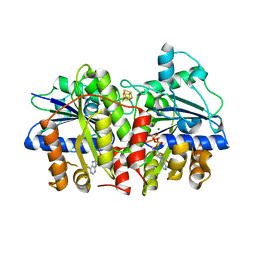 | | MgADP-bound Fe protein of Vanadium nitrogenase from Azotobacter vinelandii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Rohde, M, Gerhardt, S, Einsle, O. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of VnfH, the iron protein component of vanadium nitrogenase.
J. Biol. Inorg. Chem., 23, 2018
|
|
1ZS5
 
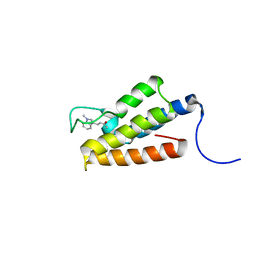 | | Structure-based evaluation of selective and non-selective small molecules that block HIV-1 TAT and PCAF association | | Descriptor: | (3E)-4-(1-METHYL-1H-INDOL-3-YL)BUT-3-EN-2-ONE, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Godbole, S, Muller, M, Yan, S, Sanchez, R, Zhou, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure-based evaluation of selective nad non-selective small molecules that block hiv-1 tat and pcaf association
TO BE PUBLISHED
|
|
6QAG
 
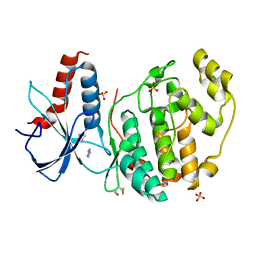 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-1,2,3-triazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
1RC9
 
 | |
6QAZ
 
 | | Crystal structure of gp41-1 intein | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Mikula, K.M, Beyer, H.M, Li, M, Wlodawer, A, Iwai, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the naturally split gp41-1 intein guides the engineering of orthogonal split inteins from cis-splicing inteins.
Febs J., 287, 2020
|
|
6QA1
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, pyridin-2-amine | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
1GXE
 
 | | Central domain of cardiac myosin binding protein C | | Descriptor: | MYOSIN BINDING PROTEIN C, CARDIAC-TYPE | | Authors: | Pfuhl, M. | | Deposit date: | 2002-04-03 | | Release date: | 2003-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Stability and Dynamics of the Central Domain of Cardiac Myosin Binding Protein C (Mybp-C): Implications for Multidomain Assembly and Causes for Cardiomyopathy
J.Mol.Biol., 329, 2003
|
|
6QD2
 
 | | MloK1 model from single particle analysis of 2D crystals, class 6 (intermediate compact conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
4P4P
 
 | | Crystal structure of Leishmania infantum polymerase beta: Nick complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Mejia, E, Burak, M, Alonso, A, Larraga, V, Kunkel, T, Bebenek, K, Garcia-Diaz, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2973 Å) | | Cite: | Structures of the Leishmania infantum polymerase beta.
DNA Repair (Amst.), 18, 2014
|
|
2RCT
 
 | | Crystal structure of human holo cellular retinol-binding protein II (CRBP-II) | | Descriptor: | L(+)-TARTARIC ACID, RETINOL, Retinol-binding protein II, ... | | Authors: | Monaco, H.L, Capaldi, S, Perduca, M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human cellular retinol-binding protein II to 1.2 A resolution.
Proteins, 70, 2007
|
|
6T21
 
 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5mCGA target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*TP*(5CM)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5CM)P*GP*AP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-10-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
6T2R
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | Beta-glycosidase, SULFATE ION | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
6QBN
 
 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200D | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Carivenc, C, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200A
To Be Published
|
|
4JGW
 
 | |
4P6R
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with tyrosine in the active site | | Descriptor: | TYROSINE, Tyrosinase, ZINC ION | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
1RGV
 
 | | Crystal Structure of the Ferredoxin from Thauera aromatica | | Descriptor: | IRON/SULFUR CLUSTER, ferredoxin | | Authors: | Unciuleac, M, Boll, M, Warkentin, E, Ermler, U. | | Deposit date: | 2003-11-13 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallization of 4-hydroxybenzoyl-CoA reductase and the structure of its electron donor ferredoxin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RW9
 
 | | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | PHOSPHATE ION, SODIUM ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Linhardt, R.J, Miyazono, H, Kyogashima, M, Kaneko, T, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
6OLK
 
 | |
1R68
 
 | | Role of the amino sugar in DNA binding of disaccharide anthracyclines: crystal structure of MAR70/d(CGATCG) complex | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Temperini, C, Cirilli, M, Aschi, M, Ughetto, G. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Role of the amino sugar in DNA binding of disaccharide anthracyclines: crystal structure of the complex MAR70/d(CGATCG).
BIOORG.MED.CHEM., 13, 2005
|
|
