6DNO
 
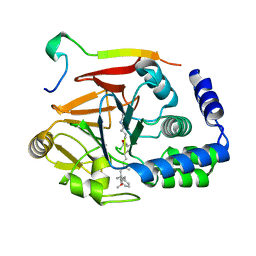 | | Crystal structure of Protein Phosphatase 1 (PP1) bound to the muscle glycogen-targeting subunit (Gm) | | Descriptor: | Microcystin-LR, Protein phosphatase 1 regulatory subunit 3A, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of the substrate recruitment mechanism of the muscle glycogen protein phosphatase 1 holoenzyme.
Sci Adv, 4, 2018
|
|
8TO5
 
 | | Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
5VM9
 
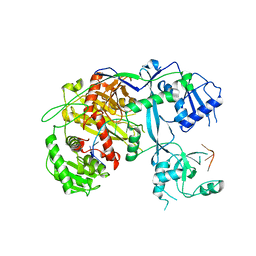 | | Human Argonaute3 bound to guide RNA | | Descriptor: | Protein argonaute-3, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3'), RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | Authors: | Park, M.S, Nakanishi, K. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Human Argonaute3 has slicer activity.
Nucleic Acids Res., 45, 2017
|
|
6AI2
 
 | |
6WY8
 
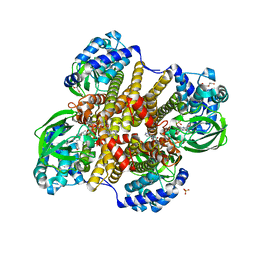 | | Tcur3481-Tcur3483 steroid ACAD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acyl-CoA dehydrogenase domain protein Tcur3481, Acyl-CoA dehydrogenase domain protein Tcur3483, ... | | Authors: | Kimber, M.S, Stirling, A.J, Seah, S.Y.K. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Key Glycine in Bacterial Steroid-Degrading Acyl-CoA Dehydrogenases Allows Flavin-Ring Repositioning and Modulates Substrate Side Chain Specificity.
Biochemistry, 59, 2020
|
|
8TRO
 
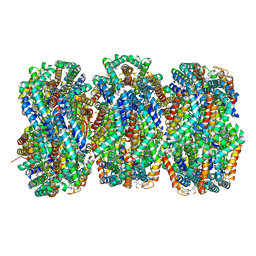 | | Rod from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8CV0
 
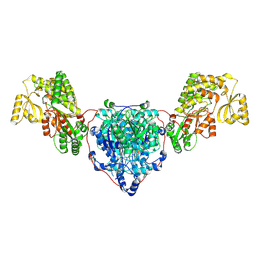 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5ZRO
 
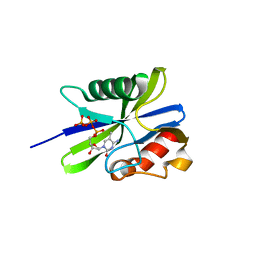 | | M. smegmatis antimutator protein MutT2 in complex with 5mdCTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
8TO2
 
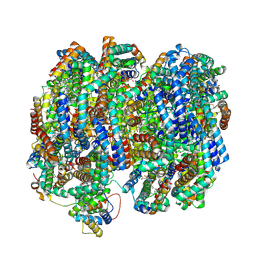 | | Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
6AH5
 
 | | Structure of human P2X3 receptor in complex with ATP and Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (3.819 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
6AI1
 
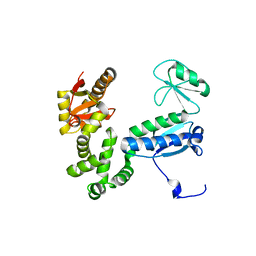 | |
8CUZ
 
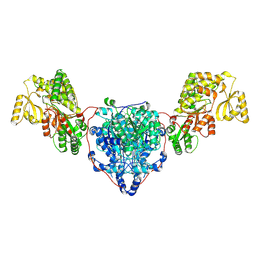 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV1
 
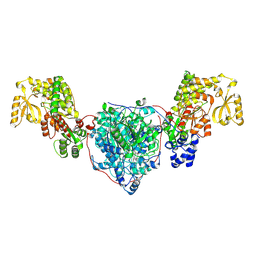 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1A5H
 
 | |
6R5W
 
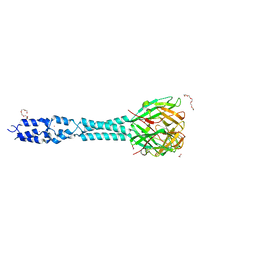 | | Crystal structure of the receptor binding protein (gp15) of Listeria phage PSA | | Descriptor: | ACETATE ION, CADMIUM ION, Gp15 protein, ... | | Authors: | Dunne, M, Ernst, P, Pluckthun, A, Loessner, M.J, Kilcher, S. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins.
Cell Rep, 29, 2019
|
|
3EB0
 
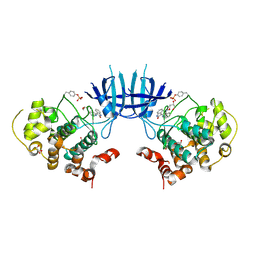 | | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, GLYCEROL, Putative uncharacterized protein | | Authors: | Wernimont, A.K, Fedorov, O, Lam, A, Ali, A, Zhao, Y, Lew, J, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804
TO BE PUBLISHED
|
|
3ED1
 
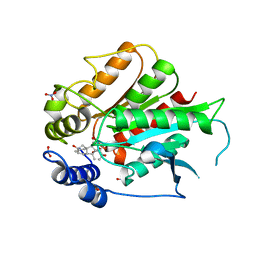 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
6RD0
 
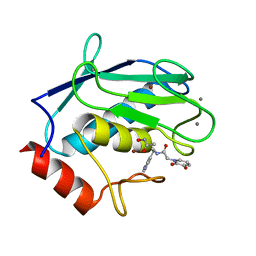 | | Human MMP12 catalytic domain in complex with AP280 | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Calderone, V, Fragai, M, Luchinat, C. | | Deposit date: | 2019-04-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of zinc-binding groups for the design of inhibitors for the oxytocinase subfamily of M1 aminopeptidases.
Bioorg.Med.Chem., 27, 2019
|
|
3DKX
 
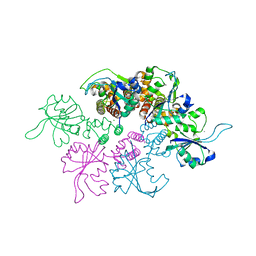 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), trigonal form, to 2.7 Ang resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
8EJN
 
 | | Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Franzen, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Comparative study of the binding and activation of 2,4-dichlorophenol by dehaloperoxidase A and B.
J.Inorg.Biochem., 247, 2023
|
|
3DKY
 
 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
8TAT
 
 | | CRYSTAL STRUCTURE OF R9A SPIN LABELED T4 LYSOZYME MUTANT K65R9A/R76R9A | | Descriptor: | Endolysin, methyl 1-hydroxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrole-3-carboxylate, radical | | Authors: | Chen, M, Hubbell, W.L, Cascio, D. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Highly Ordered Nitroxide Side Chain for Distance Mapping and Monitoring Slow Structural Fluctuations in Proteins.
Appl.Magn.Reson., 55, 2024
|
|
6R5G
 
 | |
5YVS
 
 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NADP | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
7YI7
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-[4-(trifluoromethyl)phenyl]ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
