6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7UAX
 
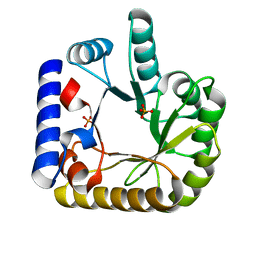 | | The crystal structure of the K36A/K38A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
5Z3F
 
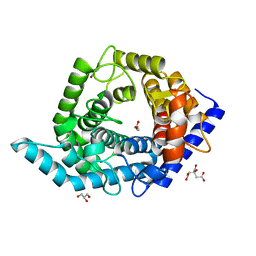 | | Glycosidase E335A in complex with glucose | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7UAT
 
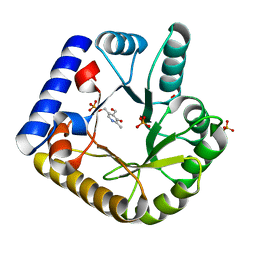 | | The crystal structure of the K36A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB8
 
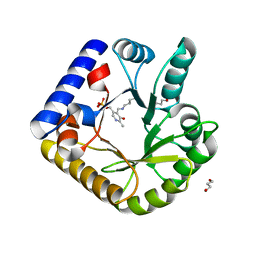 | | The crystal structure of the K38A/K137A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | 1,4-BUTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAU
 
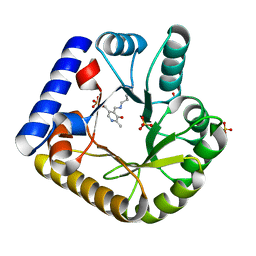 | | The crystal structure of the K137A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBQ
 
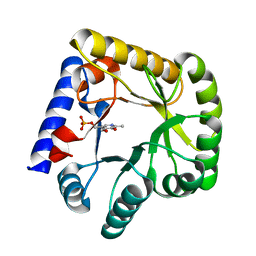 | | The crystal structure of the wild-type of E. coli YGGS in complex with PNP | | Descriptor: | PYRIDOXINE-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBP
 
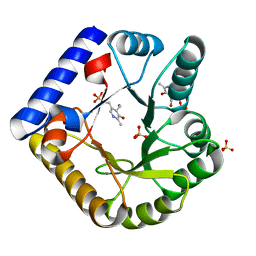 | | The crystal structure of the K36A/K137A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
6QX5
 
 | | Crystal structure of T7 bacteriophage portal protein, 12mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6HS6
 
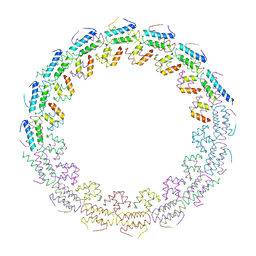 | | C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-09-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
7PAY
 
 | | Structure of the human heterotetrameric cis-prenyltransferase complex in complex with magnesium and GGsPP | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, MAGNESIUM ION, ... | | Authors: | Giladi, M, Lisnyansky Bar-El, M, Haitin, Y. | | Deposit date: | 2021-07-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for long-chain isoprenoid synthesis by cis -prenyltransferases.
Sci Adv, 8, 2022
|
|
7UB4
 
 | | The crystal structure of the K36A/K38A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
4UMY
 
 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McLean, L, Zhang, Y, Mathieu, M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
6QW3
 
 | | Calcium-bound gelsolin domain 2 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Scalone, E, Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of gelsolin domain 2 in complex with the physiological calcium ion.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
7PB1
 
 | | Structure of the human heterotetrameric cis-prenyltransferase complex in complex with magnesium, GGPP and IsPP | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, ... | | Authors: | Giladi, M, Lisnyansky Bar-El, M, Haitin, Y. | | Deposit date: | 2021-07-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for long-chain isoprenoid synthesis by cis -prenyltransferases.
Sci Adv, 8, 2022
|
|
7PAX
 
 | | Structure of the human heterotetrameric cis-prenyltransferase complex in complex with magnesium, FsPP and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, ... | | Authors: | Giladi, M, Lisnyansky Bar-El, M, Haitin, Y. | | Deposit date: | 2021-07-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for long-chain isoprenoid synthesis by cis -prenyltransferases.
Sci Adv, 8, 2022
|
|
7PB0
 
 | | Structure of the human heterotetrameric cis-prenyltransferase complex in complex with magnesium, GGsPP and IsPP | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, ... | | Authors: | Giladi, M, Lisnyansky Bar-El, M, Haitin, Y. | | Deposit date: | 2021-07-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for long-chain isoprenoid synthesis by cis -prenyltransferases.
Sci Adv, 8, 2022
|
|
1ABQ
 
 | |
4UF2
 
 | | Deerpox virus DPV022 in complex with Bax BH3 | | Descriptor: | Antiapoptotic membrane protein, Apoptosis regulator BAX | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UF3
 
 | | Deerpox virus DPV022 in complex with Bim BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2-like protein 11 | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7UTF
 
 | | Structure-Function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol | | Descriptor: | CITRATE ANION, Putative oxidoreductase, aryl-alcohol dehydrogenase like protein, ... | | Authors: | Abraham, N, Schroeter, K.L, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol.
Sci Rep, 12, 2022
|
|
4UPV
 
 | | Low X-ray dose structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
7X93
 
 | |
