3HKO
 
 | | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40 | | Descriptor: | Calcium/calmodulin-dependent protein kinase with a kinase domain and 2 calmodulin-like EF hands, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Wasney, G, Vedadi, M, MacKenzie, F, Kozieradzki, I, Cossar, D, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-25 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40
To be Published
|
|
3ZOY
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (hexagonal space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZVJ
 
 | | Crystal structure of high molecular weight (HMW) form of Peroxiredoxin I from Schistosoma mansoni | | Descriptor: | THIOREDOXIN PEROXIDASE | | Authors: | Saccoccia, F, Angelucci, F, Bellelli, A, Boumis, G, Brunori, M, Miele, A.E. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Moonlighting by Different Stressors: Crystal Structure of the Chaperone Species of a 2-Cys Peroxiredoxin.
Structure, 20, 2012
|
|
4AIT
 
 | |
3FS3
 
 | | Crystal structure of malaria parasite Nucleosome Assembly Protein (NAP) | | Descriptor: | Nucleosome assembly protein 1, putative | | Authors: | Gill, J, Yogavel, M, Kumar, A, Belrhali, H, Sharma, A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of malaria parasite nucleosome assembly protein: distinct modes of protein localization and histone recognition.
J.Biol.Chem., 284, 2009
|
|
4WWU
 
 | | Structure of Mex67:Mtr2 | | Descriptor: | ZINC ION, mRNA export factor MEX67, mRNA transport regulator MTR2 | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-12 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Domain organization within the nuclear export factor Mex67:Mtr2 generates an extended mRNA binding surface.
Nucleic Acids Res., 43, 2015
|
|
4AQF
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
476D
 
 | | CALCIUM FORM OF B-DNA UNDECAMER GCGAATTCGCG | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Tereshko, V, Egli, M. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing.
J.Mol.Biol., 291, 1999
|
|
4AHY
 
 | | Flo5A cocrystallized with 3 mM GdAc3 | | Descriptor: | CHLORIDE ION, FLOCCULATION PROTEIN FLO5, GADOLINIUM ATOM | | Authors: | Veelders, M, Essen, L.-O. | | Deposit date: | 2012-02-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex Gadolinium-Oxo Clusters Formed Along Concave Protein Surfaces.
Chembiochem, 13, 2012
|
|
4BVE
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4A00
 
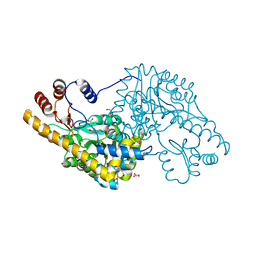 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
4A4K
 
 | |
401D
 
 | |
4A8C
 
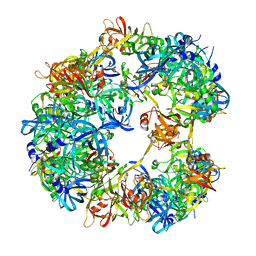 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AA2
 
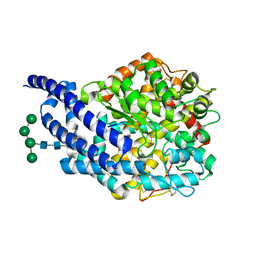 | | Crystal structure of ANCE in complex with bradykinin potentiating peptide b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ-POTENTIATING PEPTIDE B, ... | | Authors: | Isaac, R.E, Akif, M, Schwager, S.L.U, Masuyer, G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-11-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
466D
 
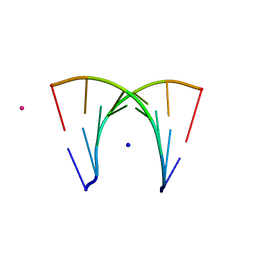 | | DISORDER AND TWIN REFINEMENT OF RNA HEPTAMER DOUBLE HELIX | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3'), SODIUM ION, ... | | Authors: | Mueller, U, Muller, Y.A, Herbst-Irmer, R, Sprinzl, M, Heinemann, U. | | Deposit date: | 1999-04-14 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Disorder and twin refinement of RNA heptamer double helices.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4A16
 
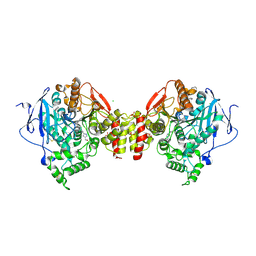 | | Structure of mouse Acetylcholinesterase complex with Huprine derivative | | Descriptor: | (1-{4-[(7S,11S)-12-AMINO-3-CHLORO-6,7,10,11-TETRAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-9-YL]BUTYL}-1H-1,2,3-TRIAZOL-4-YL)METHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Carletti, E, Colletier, J.P, Nachon, F, Weik, M, Ronco, C, Jean, L, Renard, P.Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Huprine Derivatives as Sub-Nanomolar Human Acetylcholinesterase Inhibitors: From Rational Design to Validation by X-Ray Crystallography.
Chemmedchem, 7, 2012
|
|
4AFL
 
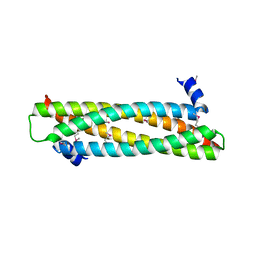 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
1H4S
 
 | | Prolyl-tRNA synthetase from Thermus thermophilus complexed with tRNApro(CGG) and a prolyl-adenylate analogue | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, PROLYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Succession of Substrate Induced Conformational Changes Ensures the Amino Acid Specificity of Thermus Thermophilus Prolyl-tRNA Synthetase: Comparison with Histidyl-tRNA Synthetase
J.Mol.Biol., 309, 2001
|
|
4AQG
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
1QI9
 
 | | X-RAY SIRAS STRUCTURE DETERMINATION OF A VANADIUM-DEPENDENT HALOPEROXIDASE FROM ASCOPHYLLUM NODOSUM AT 2.0 A RESOLUTION | | Descriptor: | VANADATE ION, Vanadium-dependent bromoperoxidase | | Authors: | Weyand, M, Hecht, H.-J, Kiess, M, Liaud, M.F, Vilter, H, Schomburg, D. | | Deposit date: | 1999-06-10 | | Release date: | 2000-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution.
J.Mol.Biol., 293, 1999
|
|
4APC
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
3ESZ
 
 | | K2AK3A Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
4AB7
 
 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae complexed with its substrate N- acetylglutamate | | Descriptor: | N-ACETYL-L-GLUTAMATE, PROTEIN ARG5,6, MITOCHONDRIAL | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
4AOF
 
 | | Selective small molecule inhibitor discovered by chemoproteomic assay platform reveals regulation of Th17 cell differentiation by PI3Kgamma | | Descriptor: | N-[6-(5-methylsulfonylpyridin-3-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]ethanamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Bergamini, G, Bell, K, Shimamura, S, Werner, T, Cansfield, A, Muller, K, Perrin, J, Rau, C, Ellard, K, Hopf, C, Doce, C, Leggate, D, Mangano, R, Mathieson, T, OMahony, A, Plavec, I, Rharbaoui, F, Reinhard, F, Savitski, M.M, Ramsden, N, Hirsch, E, Drewes, G, Rausch, O, Bantscheff, M, Neubauer, G. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Selective Inhibitor Reveals Pi3Kgamma Dependence of T(H)17 Cell Differentiation.
Nat.Chem.Biol., 8, 2012
|
|
