2KO1
 
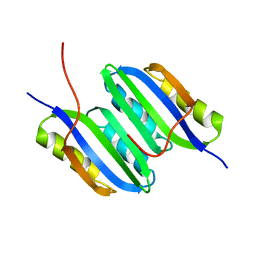 | | Solution NMR structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum. Northeast Structural Genomics Consortium Target CtR148A | | Descriptor: | GTP pyrophosphokinase | | Authors: | Eletsky, A, Garcia, E, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum
To be Published
|
|
2KKF
 
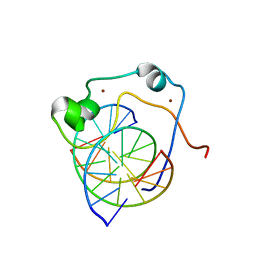 | | Solution structure of MLL CXXC domain in complex with palindromic CPG DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*CP*GP*CP*AP*GP*GP*G)-3', Histone-lysine N-methyltransferase HRX, ZINC ION | | Authors: | Cierpicki, T, Riesbeck, J.E, Grembecka, J.E, Lukasik, S.M, Popovic, R, Omonkowska, M, Shultis, D.S, Zeleznik-Le, N.J, Bushweller, J.H. | | Deposit date: | 2009-06-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1OGI
 
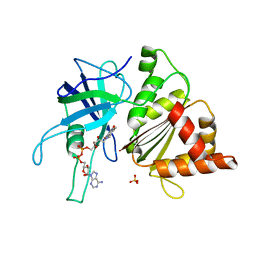 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY AND ALA 160 REPLACED BY THR (T155G-A160T) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Julvez, M.M, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Involvement of the Pyrophosphate and the 2'-Phosphate Binding Regions of Ferredoxin-Nadp+ Reductase in Coenzyme Specificity.
J.Biol.Chem., 278, 2003
|
|
4DW6
 
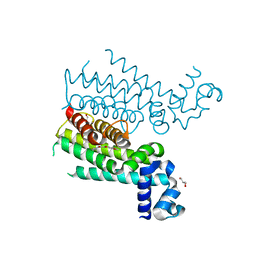 | | Novel N-phenyl-phenoxyacetamide derivatives as potential EthR inhibitors and ethionamide boosters. Discovery and optimization using High-Throughput Synthesis. | | Descriptor: | AMMONIUM ION, GLYCEROL, HTH-type transcriptional regulator EthR, ... | | Authors: | Flipo, M, Willand, N, Lecat-Guillet, N, Hounsou, C, Desroses, M, Leroux, F, Lens, Z, Villeret, V, Wohlkonig, A, Wintjens, R, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, Baulard, A.R, Deprez, B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel N-phenylphenoxyacetamide derivatives as EthR inhibitors and ethionamide boosters by combining high-throughput screening and synthesis.
J.Med.Chem., 55, 2012
|
|
2KKP
 
 | | Solution NMR structure of the phage integrase SAM-like Domain from Moth 1796 from Moorella thermoacetica. Northeast Structural Genomics Consortium Target MtR39K (residues 64-171). | | Descriptor: | Phage integrase | | Authors: | Ramelot, T.A, Wang, H, Ciccosanti, C, Jang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phage integrase SAM-like Domain from Moth 1796
from Moorella thermoacetica. Northeast
Structural Genomics Consortium Target MtR39K (residues 64-171).
To be Published
|
|
2KL4
 
 | | NMR structure of the protein NB7804A | | Descriptor: | BH2032 protein | | Authors: | Mohanty, B, Geralt, M, Augustyniak, W, Serrano, P, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein NB7804A from Bacillus Halodurans
To be Published
|
|
2KQQ
 
 | | NMR structure of human insulin mutant gly-b8-d-ala, his-b10-asp, pro-b28-lys, lys-b29-pro, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Wan, Z.L, Zhao, M, Jia, W, Huang, K, Weiss, M.A. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-24 | | Last modified: | 2021-10-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of an Turn-Stabilize But Ina Insulin Analog.
To be Published
|
|
2KLU
 
 | |
4BLJ
 
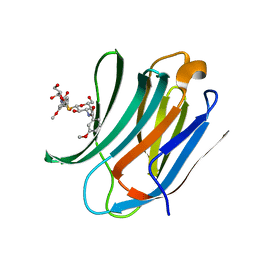 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 2 | | Descriptor: | Bis-(3-deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
2KRB
 
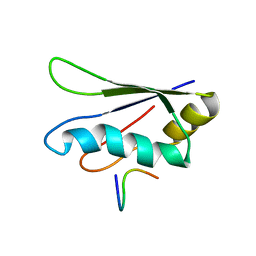 | | Solution structure of EIF3B-RRM bound to EIF3J peptide | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit J | | Authors: | Elantak, L, Wagner, S, Herrmannova, A, Janoskova, M, Rutkai, E, Lukavsky, P.J, Valasek, L. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The indispensable N-terminal half of eIF3j/HCR1 co-operates with
its structurally conserved binding partner eIF3b/PRT1-RRM and eIF1A in
stringent AUG selection
To be Published
|
|
4CLS
 
 | | Crystal structure of human soluble Adenylyl Cyclase with Pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CG0
 
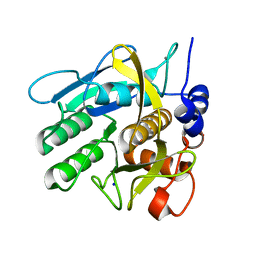 | | Savinase crystal structures for combined single crystal diffraction and powder diffraction analysis | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CS8
 
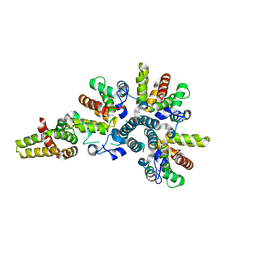 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 2 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
2KAE
 
 | | data-driven model of MED1:DNA complex | | Descriptor: | 5'-D(*DCP*DGP*DGP*DAP*DAP*DAP*DAP*DGP*DTP*DAP*DTP*DAP*DCP*DTP*DTP*DTP*DTP*DCP*DCP*DG)-3', GATA-type transcription factor, ZINC ION | | Authors: | Lowry, J.A, Gamsjaeger, R, Thong, S, Hung, W, Kwan, A.H, Broitman-Maduro, G, Matthews, J.M, Maduro, M, Mackay, J.P. | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of MED-1 Reveals Unexpected Diversity in the Mechanism of DNA Recognition by GATA-type Zinc Finger Domains.
J.Biol.Chem., 284, 2009
|
|
2K60
 
 | | NMR structure of calcium-loaded STIM1 EF-SAM | | Descriptor: | CALCIUM ION, PROTEIN (Stromal interaction molecule 1) | | Authors: | Stathopulos, P.B, Ikura, M. | | Deposit date: | 2008-07-02 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry.
Cell(Cambridge,Mass.), 135, 2008
|
|
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4CHK
 
 | | Crystal Structure of the ARF5 oligomerization domain | | Descriptor: | AUXIN RESPONSE FACTOR 5 | | Authors: | Nanao, M.H, Mazzoleni, M, Thevenon, E, Brunoud, G, Vernoux, T, Parcy, F, Dumas, R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for Oligomerisation of Auxin Transcriptional Regulators
Nat.Commun., 5, 2014
|
|
4COP
 
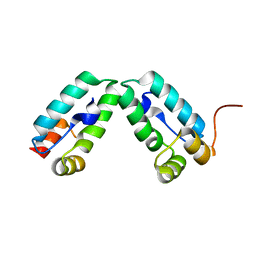 | | HIV-1 capsid C-terminal domain mutant (Y169S) | | Descriptor: | CAPSID PROTEIN P24 | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CS7
 
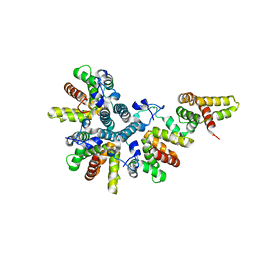 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 1 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4CU7
 
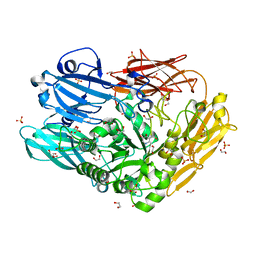 | | Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA | | Descriptor: | 1,2-ETHANEDIOL, BETA-GALACTOSIDASE, D-galacto-isofagomine, ... | | Authors: | Singh, A.K, Pluvinage, B, Higgins, M.A, Dalia, A.B, Flynn, M, Lloyd, A.R, Weiser, J.N, Stubbs, K.A, Boraston, A.B, King, S.J. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unravelling the Multiple Functions of the Architecturally Intricate Streptococcus Pneumoniae Beta-Galactosidase, BgaA.
Plos Pathog., 10, 2014
|
|
4CSC
 
 | |
2PIY
 
 | |
2PIZ
 
 | |
2KCU
 
 | | NMR solution structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107 | | Descriptor: | protein CtR107 | | Authors: | Mills, J.L, Zhang, Q, Sukumaran, D.K, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107
To be Published
|
|
2PLW
 
 | | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052). | | Descriptor: | Ribosomal RNA methyltransferase, putative, S-ADENOSYLMETHIONINE, ... | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052).
To be Published
|
|
