3OCI
 
 | | Crystal structure of TBP (TATA box binding protein) | | Descriptor: | 1,2-ETHANEDIOL, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Cui, S, Wollmann, P, Moldt, M, Hopfner, K.-P. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
4I52
 
 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
3OGR
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
3OH8
 
 | | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91 | | Descriptor: | Nucleoside-diphosphate sugar epimerase (SulA family) | | Authors: | Vorobiev, S, Lew, S, Kuzin, A, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91.
To be Published
|
|
4I7M
 
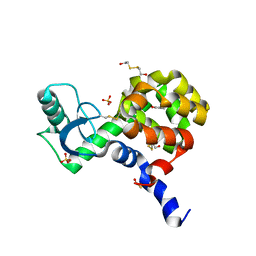 | | T4 Lysozyme L99A/M102H with 2-allylphenol bound | | Descriptor: | 2-ALLYLPHENOL, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
3OAU
 
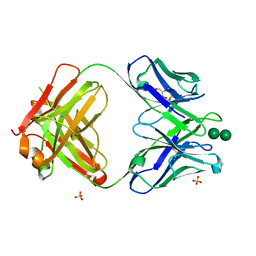 | | Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Huber, M, Wilson, I.A, Burton, D.R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged.
J.Virol., 84, 2010
|
|
4I66
 
 | |
2B8K
 
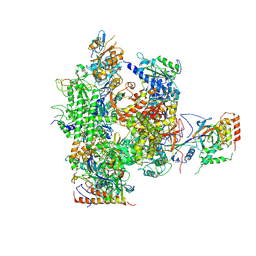 | | 12-subunit RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Meyer, P.A, Ye, P, Zhang, M, Suh, M.H, Fu, J. | | Deposit date: | 2005-10-07 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Phasing RNA Polymerase II Using Intrinsically Bound Zn Atoms: An Updated Structural Model.
Structure, 14, 2006
|
|
2BCH
 
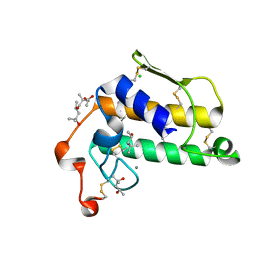 | | A possible of Second calcium ion in interfacial binding: Atomic and Medium resolution crystal structures of the quadruple mutant of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Poi, M.J, Dauter, Z, Tsai, M.D. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Suggestive evidence for the involvement of the second calcium and surface loop in interfacial binding: monoclinic and trigonal crystal structures of a quadruple mutant of phospholipase A(2).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4I6Z
 
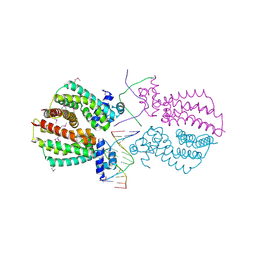 | | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide | | Descriptor: | DNA OLIGONUCLEOTIDE, Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide
To be Published
|
|
2BCR
 
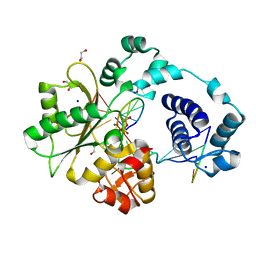 | | DNA polymerase lambda in complex with a DNA duplex containing an unpaired Damp | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*AP*TP*AP*CP*TP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
2ASC
 
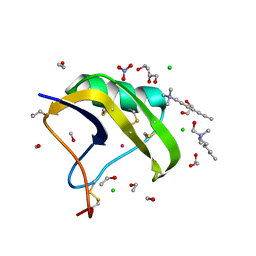 | | Scorpion toxin LQH-alpha-IT | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Kahn, R, Karbat, I, Gurevitz, M, Frolow, F. | | Deposit date: | 2005-08-23 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structures of Lqh-alpha-IT and Lqh-alpha-IT8D9D10V mutant
To be Published
|
|
4I7B
 
 | | Siah1 bound to synthetic peptide (ACE)KLRPV(ABA)MVRPTVR | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ZINC ION | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I7N
 
 | | T4 Lysozyme L99A/M102H with 1-phenyl-2-propyn-1-ol bound | | Descriptor: | (1R)-1-phenylprop-2-yn-1-ol, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
2ASV
 
 | |
3OGS
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
2AJ2
 
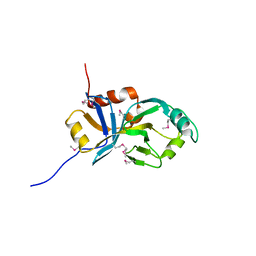 | | X-Ray Crystal Structure of Protein VC0467 from Vibrio cholerae. Northeast Structural Genomics Consortium Target VcR8. | | Descriptor: | Hypothetical UPF0301 protein VC0467 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | X-Ray structure of hypothetical protein VC0467 from Vibrio cholerae: new fold. Northeast Structural Genomics Consortium target VcR8.
To be Published
|
|
4K0C
 
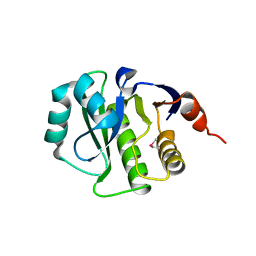 | | Crystal Structure of the computationally designed serine hydrolase. Northeast Structural Genomics Consortium (NESG) Target OR317 | | Descriptor: | designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR317
To be Published
|
|
3OGF
 
 | |
2AMP
 
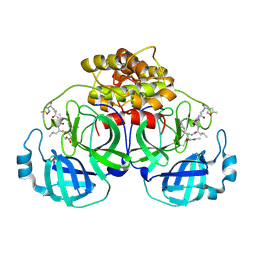 | | Crystal Structure Of Porcine Transmissible Gastroenteritis Virus Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
4KH7
 
 | | Crystal structure of a glutathione transferase family member from salmonella enterica ty2, target efi-507262, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione s-transferase family protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Salmonella enterica ty2, target efi-507262, with bound glutathione
To be published
|
|
2AQU
 
 | | Structure of HIV-1 protease bound to atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, HIV-1 Protease | | Authors: | Clemente, J.C, Coman, R.M, Thiaville, M.M, Janka, L.K, Jeung, J.A, Nukoolkarn, S, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Leelamanit, W, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of HIV-1 CRF_01 A/E protease inhibitor resistance: structural determinants for maintaining sensitivity and developing resistance to atazanavir.
Biochemistry, 45, 2006
|
|
3OJ8
 
 | | Alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain | | Descriptor: | (S)-[(2S)-6-phenoxy-1,2,3,4-tetrahydronaphthalen-2-yl](5-pyridin-2-yl-1,3-oxazol-2-yl)methanol, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2010-08-20 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain
J.Med.Chem., 54, 2011
|
|
3OK2
 
 | |
2AUJ
 
 | | Structure of Thermus aquaticus RNA polymerase beta'-subunit insert | | Descriptor: | DNA-directed RNA polymerase beta' chain | | Authors: | Chlenov, M, Masuda, S, Murakami, K.S, Nikiforov, V, Darst, S.A, Mustaev, A. | | Deposit date: | 2005-08-27 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of Lineage-specific Sequence Insertions in the Bacterial RNA Polymerase beta' Subunit
J.Mol.Biol., 353, 2005
|
|
