6L1V
 
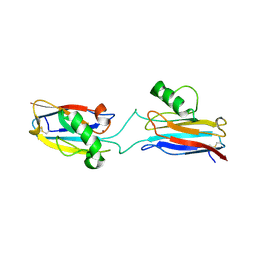 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | Descriptor: | Azurin-1, COPPER (II) ION | | Authors: | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
9B9V
 
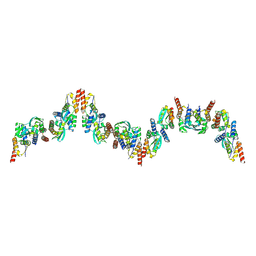 | |
6KVD
 
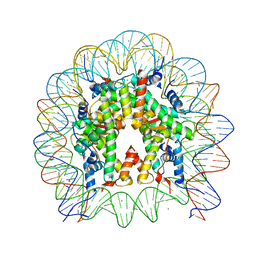 | | Crystal structure of human nucleosome containing H2A.J | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.J, ... | | Authors: | Tanaka, H, Koyama, M, Sato, S, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and structural analyses of the nucleosome containing human histone H2A.J.
J.Biochem., 167, 2020
|
|
5OJ8
 
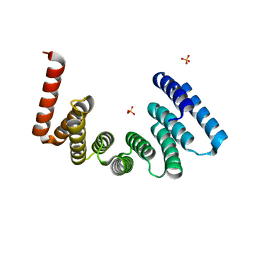 | | Crystal structure of the KLC1-TPR domain ([A1-B5] fragment) | | Descriptor: | Kinesin light chain 1, PHOSPHATE ION | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
4UVC
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Phenyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-[5-[(1S)-1-azanyl-1,3-diphenyl-propyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-4aH-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
9BF2
 
 | | MID domain of Ago2 bound to UMP | | Descriptor: | Protein argonaute-2, URIDINE-5'-MONOPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9EZX
 
 | | Vibrio cholerae DdmD apo complex | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
9BF0
 
 | | MID domain of human Argo2 bound to UTP | | Descriptor: | Protein argonaute-2, URIDINE 5'-TRIPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9FTQ
 
 | | Drosophila golgi alpha-mannosidase II (dGMII) in complex with swainsonine-configured alkyl indolizidine | | Descriptor: | Alpha-mannosidase 2, DI(HYDROXYETHYL)ETHER, ZINC ION, ... | | Authors: | Bennett, M, Koemans, T, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2024-06-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure-guided design of C3-branched swainsonine as potent and selective human Golgi alpha-mannosidase (GMII) inhibitor.
Chem.Commun.(Camb.), 60, 2024
|
|
9FTR
 
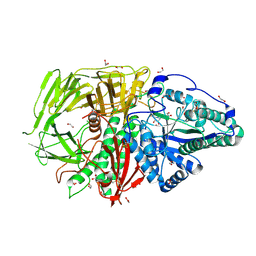 | | Drosophila golgi alpha-mannosidase II (dGMII) in complex with amide modified swainsonine-configured alkyl indolizidine | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION, ... | | Authors: | Bennett, M, Koemans, T, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2024-06-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-guided design of C3-branched swainsonine as potent and selective human Golgi alpha-mannosidase (GMII) inhibitor.
Chem.Commun.(Camb.), 60, 2024
|
|
5OJF
 
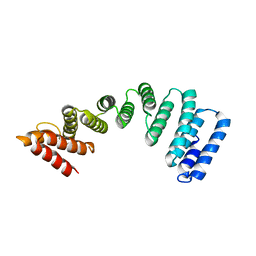 | | Crystal Structure of KLC2-TPR domain (fragment [A1-B6] | | Descriptor: | Kinesin light chain 2 | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Andreani, J, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
9ERO
 
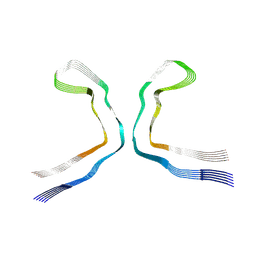 | |
7X01
 
 | | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with inhibitor FHA | | Descriptor: | (1R,2S,3S,4R,5R)-3-(6-aminopurin-9-yl)-4-fluoranyl-5-(2-hydroxyethyl)cyclopentane-1,2-diol, ZINC ION, mRNA-capping enzyme nsP1 | | Authors: | Zhang, K, Law, M.C.Y, Nguyen, T.M, Tan, Y.B, Wirawan, M, Law, Y.S, Luo, D.H. | | Deposit date: | 2022-02-20 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1.
Cell Rep, 40, 2022
|
|
9EO6
 
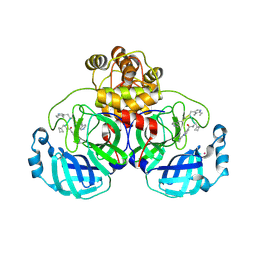 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOX
 
 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, Soluble inhibitor | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
7XL0
 
 | | Crystal structure of Vobarilizumab at 1.70 Angstrom | | Descriptor: | GLYCEROL, Nanobody Vobarilizumab, SULFATE ION | | Authors: | Caaveiro, J.M.M, Mori, C, Kinoshita, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
6L0Q
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL12, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
7XL1
 
 | | Crystal structure of chimeric 7D12-Vob nanobody at 1.65 Angstrom | | Descriptor: | Chimeric 7D12-Vob nanobody, MALONATE ION | | Authors: | Caaveiro, J.M.M, Kinoshita, S, Mori, C, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
6L2N
 
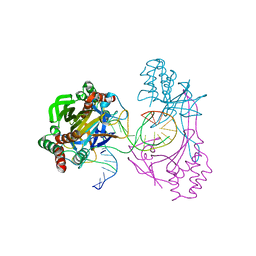 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-3bp-GTAC) complex | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*GP*TP*AP*CP*TP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*GP*A)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
9FCT
 
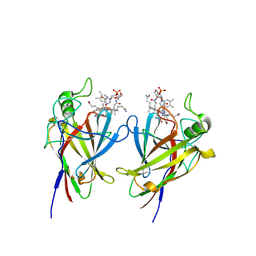 | |
9ERM
 
 | |
4WCB
 
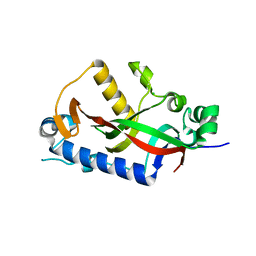 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H309Q | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDE
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T311A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
6L0P
 
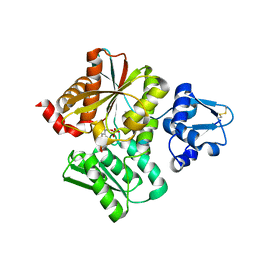 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
