2MWJ
 
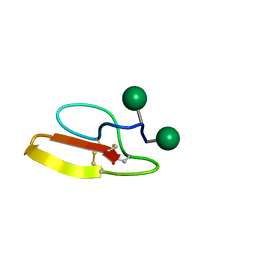 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
5UZ9
 
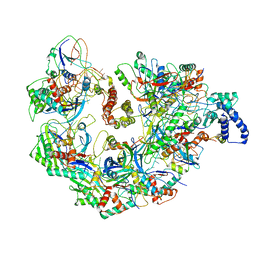 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
5UU5
 
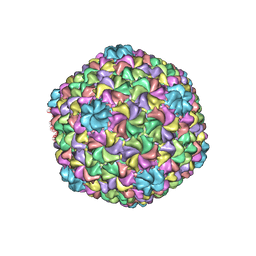 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6CP6
 
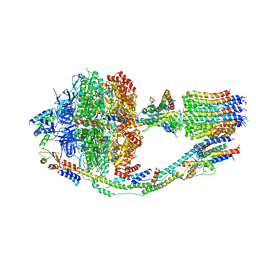 | | Monomer yeast ATP synthase (F1Fo) reconstituted in nanodisc. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
2HMX
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 MATRIX PROTEIN | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 MATRIX PROTEIN | | Authors: | Massiah, M.A, Starich, M.R, Paschall, C, Christensen, A.M, Sundquist, W.I, Summers, M.F. | | Deposit date: | 1995-09-22 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein.
J.Mol.Biol., 244, 1994
|
|
2HHB
 
 | |
6CP7
 
 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
1WAW
 
 | | Specificity and affinity of natural product cyclopentapeptide inhibitor Argadin against human chitinase | | Descriptor: | ARGADIN, CHITOTRIOSIDASE 1, GLYCEROL, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-28 | | Release date: | 2005-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1W9V
 
 | | Specificity and affinity of natural product cyclopentapeptide argifin against Aspergillus fumigatus | | Descriptor: | ARGIFIN, CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1W9U
 
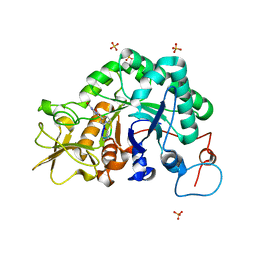 | | Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against Aspergillus fumigatus chitinase | | Descriptor: | ARGADIN, CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1W9P
 
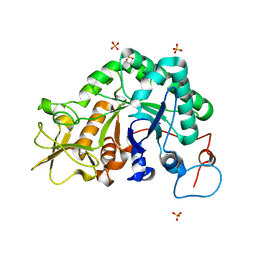 | | Specificity and affinity of natural product cyclopentapeptide inhibitors against Aspergillus fumigatus, human and bacterial chitinaseFra | | Descriptor: | CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, Van Aalten, D.M.F. | | Deposit date: | 2004-10-15 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1WB0
 
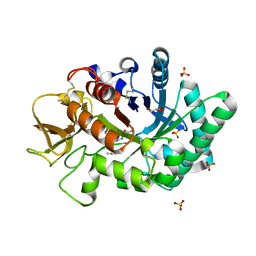 | | specificity and affinity of natural product cyclopentapeptide inhibitor Argifin against human chitinase | | Descriptor: | ARGIFIN, CHITOTRIOSIDASE 1, GLYCEROL, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, Van Aalten, D.M.F. | | Deposit date: | 2004-10-29 | | Release date: | 2005-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
3ZFV
 
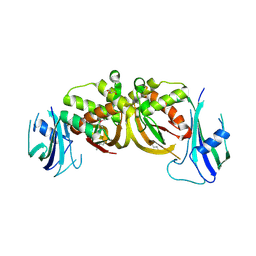 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
3ZC4
 
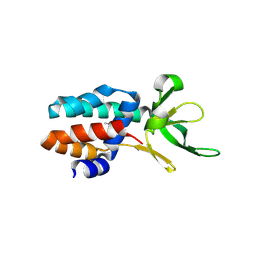 | | The structure of Csa5 from Sulfolobus solfataricus. | | Descriptor: | DI(HYDROXYETHYL)ETHER, SSO1398 | | Authors: | Reeks, J, Anderson, L, White, M.F, Naismith, J.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the Archaeal Cascade Subunit Csa5: Relating the Small Subunits of Crispr Effector Complexes.
RNA Biol., 10, 2013
|
|
3ZJ0
 
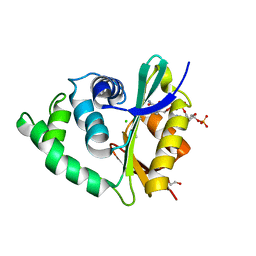 | | The human O-GlcNAcase C-terminal domain is a pseudo histone acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ACETYLTRANSFERASE, ... | | Authors: | Rao, F.V, Schuettelkopf, A.W, Dorfmueller, H.C, Ferenbach, A.T, Navratilova, I, van Aalten, D.M.F. | | Deposit date: | 2013-01-15 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Bacterial Putative Acetyltransferase Defines the Fold of the Human O-Glcnacase C-Terminal Domain.
Open Biol., 3, 2013
|
|
7BG1
 
 | |
7BDV
 
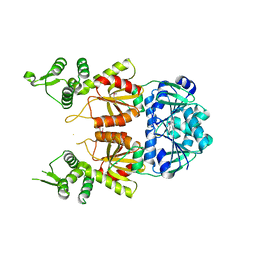 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
7EVT
 
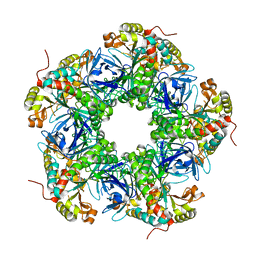 | | Crystal structure of the N-terminal degron-truncated human glutamine synthetase | | Descriptor: | Glutamine synthetase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of N-terminal degron-truncated human glutamine synthetase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6FQG
 
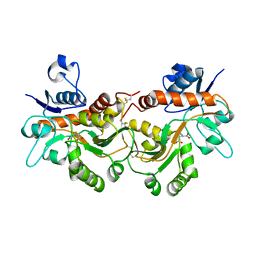 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form A) at 2.34 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34139085 Å) | | Cite: | X-ray structure of GluA2 flop G724C ligand binding core dimer bound to glutamate at 2.32 Angstroms resolution
To Be Published
|
|
5KCS
 
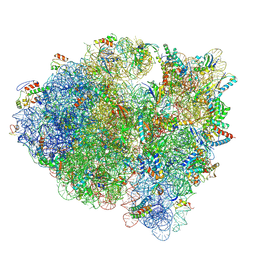 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Evernimycin, mRNA, TetM and P-site tRNA at 3.9A resolution | | Descriptor: | (2R,3R,4R,6S)-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7S,7aR)-6-{[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5S,6S)-6-({(2R,3aS,3a'R,6S,7R,7'R,7aS,7a'S)-7'-[(2,4-dihydroxy-6-methylbenzoyl)oxy]-7-hydroxyoctahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-4-hydroxy-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-3-hydroxy-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4',7-dihydroxy-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-4-{[(2R,4S,5R,6S)-5-methoxy-4,6-dimethyl-4-nitrotetrahydro-2H-pyran-2-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6IBU
 
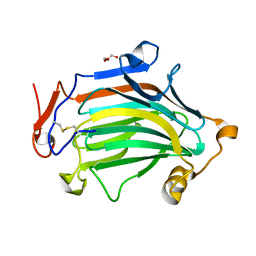 | | Apo Crh5 transglycosylase | | Descriptor: | GLYCEROL, Probable glycosidase crf1 | | Authors: | Bartual, S.G, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
6IBW
 
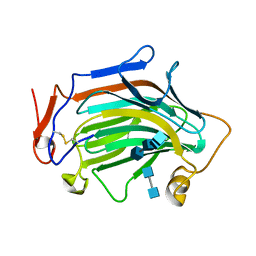 | | Crh5 transglycosylase in complex with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Probable glycosidase crf1 | | Authors: | Fang, W, Bartual, S.G, van Aalten, D.M.F. | | Deposit date: | 2018-12-01 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
1JR5
 
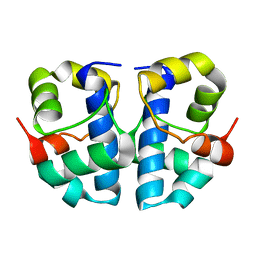 | | Solution Structure of the Anti-Sigma Factor AsiA Homodimer | | Descriptor: | 10 KDA Anti-Sigma Factor | | Authors: | Urbauer, J.L, Simeonov, M.F, Bieber Urbauer, R.J, Adelman, K, Gilmore, J.M, Brody, E.N. | | Deposit date: | 2001-08-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K1Q
 
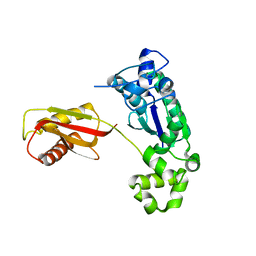 | | Crystal Structure of a DinB Family Error Prone DNA Polymerase from Sulfolobus solfataricus | | Descriptor: | DBH protein | | Authors: | Silvian, L.F, Toth, E.A, Pham, P, Goodman, M.F, Ellenberger, T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus.
Nat.Struct.Biol., 8, 2001
|
|
5KCR
 
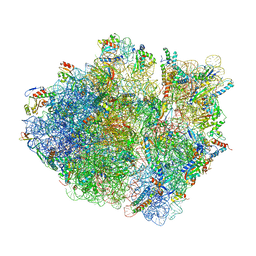 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
