8FYU
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 10mer acetylated tau peptide | | Descriptor: | ACE-SER-SER-THR-GLY-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84839141 Å) | | Cite: | Phosphorylation of a Cleaved Tau Proteoform at a Single Residue Inhibits Binding to the E3 Ubiquitin Ligase, CHIP.
Biorxiv, 2023
|
|
8FR7
 
 | | A hinge glycan regulates spike bending and impacts coronavirus infectivity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pintilie, G, Wilson, E, Chmielewski, D, Schmid, M.F, Jin, J, Chen, M, Singharoy, A, Chiu, W. | | Deposit date: | 2023-01-06 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A hinge glycan regulates spike bending and impacts coronavirus infectivity
To Be Published
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
6WN6
 
 | | Crystal structure of 3-keto-D-glucoside 4-epimerase, YcjR, from E. coli, apo form | | Descriptor: | 1,2-ETHANEDIOL, 3-keto-D-glucoside 4-epimerase, MANGANESE (II) ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Mukherjee, K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Reaction Mechanism of YcjR, an Epimerase That Facilitates the Interconversion of d-Gulosides to d-Glucosides inEscherichia coli.
Biochemistry, 59, 2020
|
|
8GRS
 
 | | human TMEM63A | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CSC1-like protein 1 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GRN
 
 | | AtOSCA1.1 extended state | | Descriptor: | Protein OSCA1, [1-MYRISTOYL-GLYCEROL-3-YL]PHOSPHONYLCHOLINE | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GSO
 
 | | AtOSCA3.1 channel extended state | | Descriptor: | CSC1-like protein ERD4 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
6WX3
 
 | | High resolution Tryptophan Synthase crystal structure from Salmonella typhimurium in complex with F9 inhibitor in the alpha-site, Cesium ion at the metal coordination site and internal aldimine form. | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution Tryptophan Synthase crystal structure from Salmonella typhimurium in complex with F9 inhibitor in the alpha-site, Cesium ion at the metal coordination site and internal aldimine form.
To be Published
|
|
8GRO
 
 | | AtOSCA3.1 contracted state | | Descriptor: | CSC1-like protein ERD4 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
6X0C
 
 | | Tryptophan Synthase mutant beta-Q114A in complex with Cesium ion at the metal coordination site and aminoacrylate and benzimidazole at the enzyme beta site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-05-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | External aldimine form of Tryptophan Synthase mutant beta-Q114A in complex with Cesium ion at the metal coordination site and benzimidazole at the enzyme beta site.
To be Published
|
|
6WDU
 
 | | The external aldimine form of the Salmonella thypi wild-type tryptophan synthase in open conformation showing multiple side chain conformations for the residue beta Q114 and sodium ion at the metal coordination site. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring. | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The external aldimine form of the Salmonella thypi wild-type tryptophan synthase in open conformation showing multiple side chain conformations for the residue beta Q114 and sodium ion at the metal coordination site. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
6WVZ
 
 | | Crystal structure of anti-MET Fab arm of amivantamab in complex with human MET | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2020-05-07 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of amivantamab (JNJ-61186372), a bispecific antibody targeting EGFR and MET.
J.Biol.Chem., 296, 2021
|
|
7OP7
 
 | | Bacteroides thetaiotaomicron mannosidase GH2 with beta-manno-configured N-alkyl cyclophellitol aziridine | | Descriptor: | (1R,2S,3R,4S,5R,6R)-5-(8-azidooctylamino)-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, BROMIDE ION, ... | | Authors: | McGregor, N.G.S, Beenakker, T.J.M, Kuo, C, Wong, C, Offen, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | Descriptor: | D-binder, L-pep1 | | Authors: | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of D-protein binder D-19437 and L-target L-Pep-1
To Be Published
|
|
7OMS
 
 | | Bs164 in complex with mannocyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McGregor, N, Beenakker, T, Kuo, C.L, Wong, C.S, Offren, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H, Davies, G.J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7OP6
 
 | | Bacteroides thetaiotaomicron mannosidase GH2 with beta-manno-configured cyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, BROMIDE ION, ... | | Authors: | McGregor, N.G.S, Beenakker, T.J.M, Kuo, C, Wong, C, Offen, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7YJ0
 
 | |
8GCK
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | Descriptor: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36823535 Å) | | Cite: | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
4KC8
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
7XTK
 
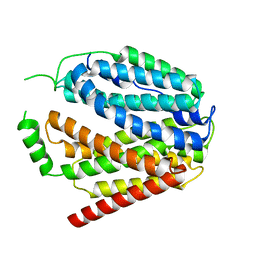 | | Cryo-EM structure of SLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, M.F, Shan, Y.Y. | | Deposit date: | 2022-05-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of human SLC19A1.
To Be Published
|
|
7OYL
 
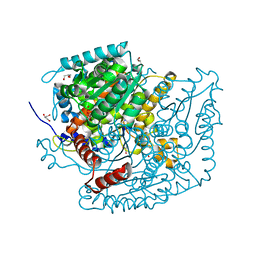 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
7XYM
 
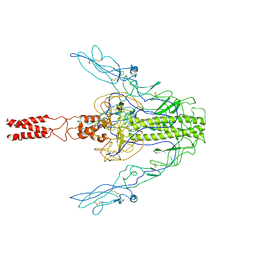 | |
4KCA
 
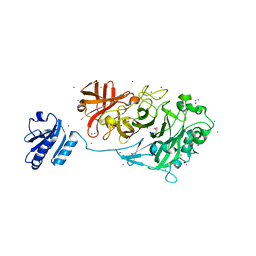 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
7P5O
 
 | |
4KCB
 
 | | Crystal Structure of Exo-1,5-alpha-L-arabinanase from Bovine Ruminal Metagenomic Library | | Descriptor: | Arabinan endo-1,5-alpha-L-arabinosidase, PHOSPHATE ION | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
