5DIY
 
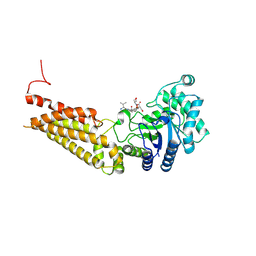 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
2V3S
 
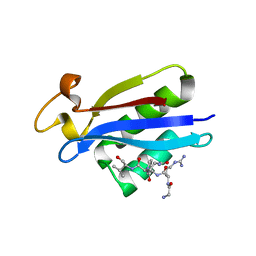 | | Structural insights into the recognition of substrates and activators by the OSR1 kinase | | Descriptor: | ACETATE ION, SERINE/THREONINE-PROTEIN KINASE OSR1, SERINE/THREONINE-PROTEIN KINASE WNK4 | | Authors: | Villa, F, Goebel, J, Rafiqi, F.H, Deak, M, Thastrup, J, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2007-06-21 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Recognition of Substrates and Activators by the Osr1 Kinase
Embo Rep., 8, 2007
|
|
4S3G
 
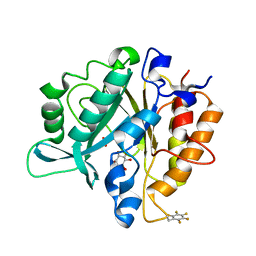 | | Structure of the F249X mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-01 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
2VXK
 
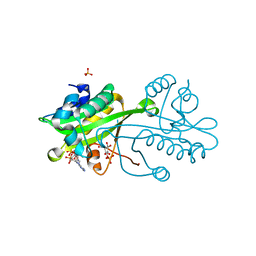 | | Structural comparison between Aspergillus fumigatus and human GNA1 | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, GLUCOSAMINE 6-PHOSPHATE ACETYLTRANSFERASE, ... | | Authors: | Hurtado-Guerrero, R, Raimi, O.G, Min, J, Zeng, H, Vallius, L, Shepherd, S, Ibrahim, A.F.M, Wu, H, Plotnikov, A.N, van Aalten, D.M.F. | | Deposit date: | 2008-07-05 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Differences between Human and Aspergillus Fumigatus D-Glucosamine-6- Phosphate N-Acetyltransferase.
Biochem.J., 415, 2008
|
|
2VXZ
 
 | | Crystal Structure of hypothetical protein PyrSV_gp04 from Pyrobaculum spherical virus | | Descriptor: | CHLORIDE ION, GLYCEROL, PYRSV_GP04 | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-07-15 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2W03
 
 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with adenosine, sulfate and citrate from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE, CITRIC ACID, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2VWI
 
 | | Structure of the OSR1 kinase, a hypertension drug target | | Descriptor: | GOLD ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Villa, F, Deak, M, Alessi, D.R, vanAalten, D.M.F. | | Deposit date: | 2008-06-25 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the Osr1 Kinase, a Hypertension Drug Target.
Proteins, 73, 2008
|
|
5DS3
 
 | | Crystal structure of constitutively active PARP-1 | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
2W3Z
 
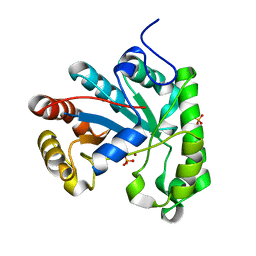 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
4UWN
 
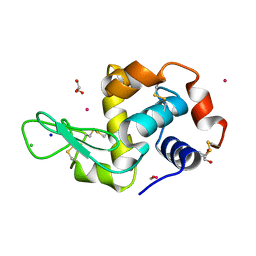 | | Lysozyme soaked with a ruthenium based CORM with a methione oxide ligand (complex 6b) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
5DOE
 
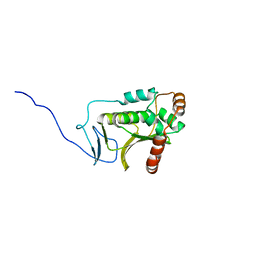 | | Crystal structure of the Human Cytomegalovirus UL53 (residues 72-292) | | Descriptor: | Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
2W04
 
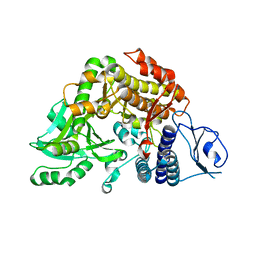 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2VUR
 
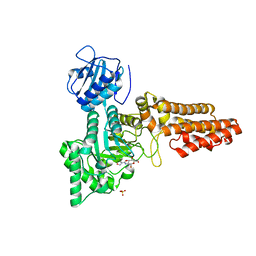 | | Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death | | Descriptor: | 2-deoxy-2-{[(2-hydroxy-1-methylhydrazino)carbonyl]amino}-beta-D-glucopyranose, O-GLCNACASE NAGJ, SULFATE ION | | Authors: | Pathak, S, Dorfmueller, H.C, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2008-05-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical Dissection of the Link between Streptozotocin, O-Glcnac, and Pancreatic Cell Death.
Chem.Biol., 15, 2008
|
|
2WB5
 
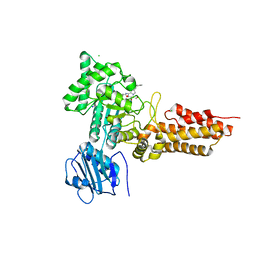 | | GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation | | Descriptor: | (5R,6R,7R,8S)-6,7-dihydroxy-5-(hydroxymethyl)-2-(2-phenylethyl)-8-(propanoylamino)-5,6,7,8-tetrahydro-1H-imidazo[1,2-a]pyridin-4-ium, CHLORIDE ION, O-GLCNACASE NAGJ, ... | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glcnacstatins are Nanomolar Inhibitors of Human O-Glcnacase Inducing Cellular Hyper-O-Glcnacylation
Biochem.J., 420, 2009
|
|
2W61
 
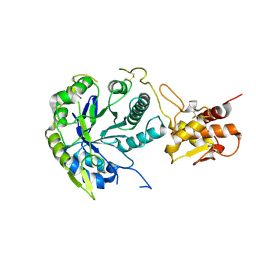 | |
4WPG
 
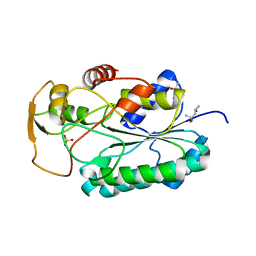 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
2W63
 
 | |
2W62
 
 | | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose | | Descriptor: | 1,4-BUTANEDIOL, GLYCOLIPID-ANCHORED SURFACE PROTEIN 2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Schuettelkopf, A.W, Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling.
J.Biol.Chem., 284, 2009
|
|
4WN4
 
 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WF1
 
 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RV3
 
 | | Crystal structure of a pentafluoro-Phe incorporated Phosphatidylinositol-specific phospholipase C (H258X)from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2014-11-24 | | Release date: | 2015-07-01 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
4WX2
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
4U78
 
 | | Octameric RNA duplex soaked in copper(II)chloride | | Descriptor: | CALCIUM ION, COPPER (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4WSL
 
 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4X3V
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | Descriptor: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Dealwis, C.G, Ahmad, M.F, Alam, I. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
