8RFK
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - single mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, Quinoprotein glucose dehydrogenase B | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RE0
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RG1
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - wild type pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
8FYU
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 10mer acetylated tau peptide | | Descriptor: | ACE-SER-SER-THR-GLY-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84839141 Å) | | Cite: | Phosphorylation of tau at a single residue inhibits binding to the E3 ubiquitin ligase, CHIP.
Nat Commun, 15, 2024
|
|
8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | Descriptor: | D-binder, L-pep1 | | Authors: | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
4PJS
 
 | | Crystal structure of designed (SeMet)-cPPR-NRE protein | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJQ
 
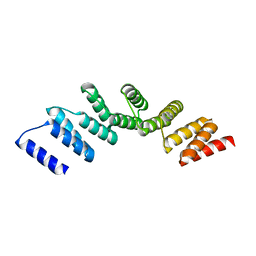 | | Crystal structure of designed cPPR-polyG protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
8RMO
 
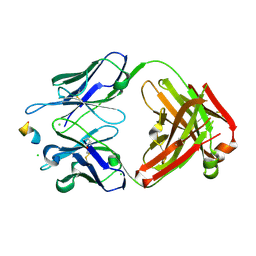 | | Crystal structure of anti-FLAG M2 Fab fragment bound to FLAG-tag peptide epitope | | Descriptor: | CHLORIDE ION, FLAG-tag, anti-FLAG M2 heavy chain, ... | | Authors: | Beugelink, J.W, Janssen, B.J.C, Pronker, M.F. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.163 Å) | | Cite: | Structural Basis for Recognition of the FLAG-tag by Anti-FLAG M2.
J.Mol.Biol., 436, 2024
|
|
8TE7
 
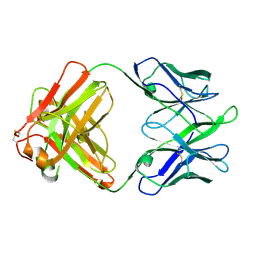 | | Structure of TRNM-f.01 | | Descriptor: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | Authors: | Bender, M.F, Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Broad and Potent HIV-1 Neutralization in Fusion Peptide-primed SHIV-boosted Macaques
To Be Published
|
|
4PJR
 
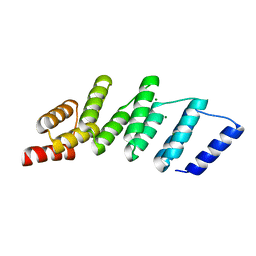 | | Crystal structure of designed cPPR-NRE protein | | Descriptor: | MAGNESIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PY6
 
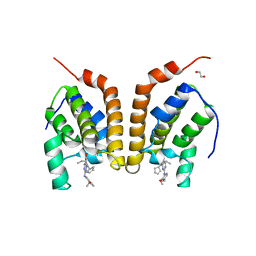 | | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain protein, ... | | Authors: | Fonseca, M, Tallant, C, Hutchinson, A, Savitsky, P, Krojer, T, Filippakopoulos, P, Loppnau, P, Brennan, P.E, von Delft, F, Dong, A, Josling, G.A, Duffy, M.F, Arrowsmith, C.H, Bountra, C, Hui, R, Knapp, S, Wernimont, A.K, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum
To be Published
|
|
4Q9B
 
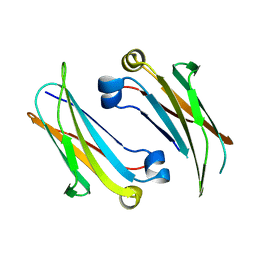 | | IgNAR antibody domain C2 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QFG
 
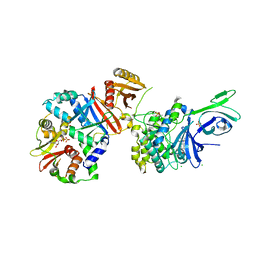 | |
4QFR
 
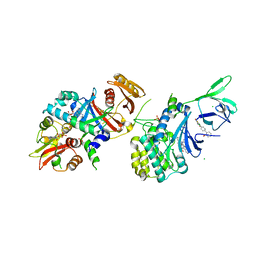 | | Structure of AMPK in complex with Cl-A769662 activator and STAUROSPORINE inhibitor | | Descriptor: | 2-chloro-4-hydroxy-3-(2'-hydroxybiphenyl-4-yl)-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4Q0C
 
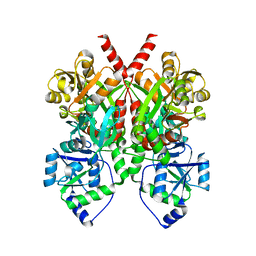 | | 3.1 A resolution crystal structure of the B. pertussis BvgS periplasmic domain | | Descriptor: | Virulence sensor protein BvgS | | Authors: | Dupre, E, Herrou, J, Lensink, M.F, Wintjens, R, Lebedev, A, Crosson, S, Villeret, V, Locht, C, Antoine, R, Jacob-Dubuisson, F. | | Deposit date: | 2014-04-01 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS.
Plos Pathog., 11, 2015
|
|
4Q9C
 
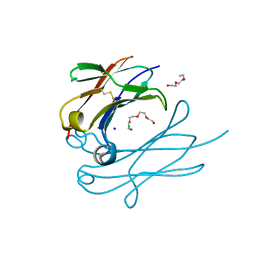 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5JJ4
 
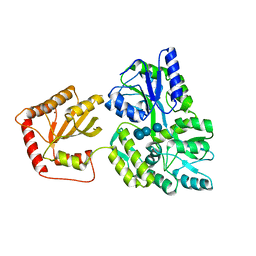 | | Crystal Structure of a Variant Human Activation-induced Deoxycytidine Deaminase as an MBP fusion protein | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Single-stranded DNA cytosine deaminase, ZINC ION, ... | | Authors: | Pedersen, L.C, Goodman, M.F, Pham, P, Afif, S.A. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural analysis of the activation-induced deoxycytidine deaminase required in immunoglobulin diversification.
DNA Repair (Amst.), 43, 2016
|
|
7PPR
 
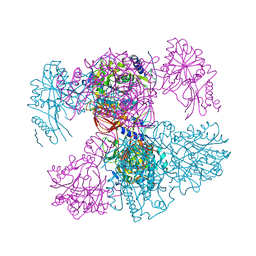 | |
7PJC
 
 | |
7QQK
 
 | | TIR-SAVED effector bound to cA3 | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), TIR_SAVED fusion protein | | Authors: | Spagnolo, L, White, M.F, Hogrel, G, Guild, A. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic nucleotide-induced helical structure activates a TIR immune effector.
Nature, 608, 2022
|
|
5KCS
 
 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Evernimycin, mRNA, TetM and P-site tRNA at 3.9A resolution | | Descriptor: | (2R,3R,4R,6S)-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7S,7aR)-6-{[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5S,6S)-6-({(2R,3aS,3a'R,6S,7R,7' R,7aS,7a'S)-7'-[(2,4-dihydroxy-6-methylbenzoyl)oxy]-7-hydroxyoctahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}ox y)-4-hydroxy-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-3-hydroxy-5-methoxy-6-methyltetrahydro-2H-pyran-4- yl]oxy}-4',7-dihydroxy-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-4-{[(2R,4S,5R, 6S)-5-methoxy-4,6-dimethyl-4-nitrotetrahydro-2H-pyran-2-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KCR
 
 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7QOV
 
 | | The wild type nitrile hydratase from Geobacillus pallidus | | Descriptor: | CHLORIDE ION, COBALT (III) ION, Nitrile hydratase, ... | | Authors: | Van Wyk, J.C, Cowan, D.A, Danson, M.J, Tsekoa, T.L, Sayed, M.F, Sewell, B.T. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering enhanced thermostability into the Geobacillus pallidus nitrile hydratase.
Curr Res Struct Biol, 4, 2022
|
|
7QOU
 
 | | A mutant of the nitrile hydratase from Geobacillus pallidus having enhanced thermostability | | Descriptor: | CHLORIDE ION, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Van Wyk, J.C, Cowan, D.A, Danson, M.J, Tsekoa, T.L, Sayed, M.F, Sewell, B.T. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering enhanced thermostability into the Geobacillus pallidus nitrile hydratase.
Curr Res Struct Biol, 4, 2022
|
|
