7X7M
 
 | | Lumazine Synthase from Aquifex aeolicus | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sobhy, M.A, Hamdan, S.M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-electron structures of the extreme thermostable enzymes Sulfur Oxygenase Reductase and Lumazine Synthase.
Plos One, 17, 2022
|
|
7XKX
 
 | | Crystal structure of Tpn2 | | Descriptor: | SQHop_cyclase_C domain-containing protein | | Authors: | Chang, C.Y, Stowell, E.A, Lin, Y.L, Ehrenberger, M.A, Rudolf, J.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure-guided product determination of the bacterial type II diterpene synthase Tpn2.
Commun Chem, 5, 2022
|
|
4TLN
 
 | |
7Y6M
 
 | |
4RXY
 
 | |
7Z4S
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with cyclic peptide GM4 including unnatural amino acids. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Miura, T, Malla, T, Lukacik, L, Strain-Damerell, C.M, Tumber, A, Brewitz, L, McDonough, M.A, Salah, E, Terasaka, N, Katoh, T, Kawamura, A, Schofield, C.J, Suga, H, Walsh, M.A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In vitro selection of macrocyclic peptide inhibitors containing cyclic gamma 2,4 -amino acids targeting the SARS-CoV-2 main protease.
Nat.Chem., 15, 2023
|
|
6V99
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase- S72D in the presence of sulfate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V96
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72E | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Hussien, R, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V9A
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72D | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
1AYN
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
6HMX
 
 | |
9FJN
 
 | | Solution NMR structure of a peptide encompassing residues 2-19 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
9FJW
 
 | | Solution NMR structure of a peptide encompassing residues 2-36 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
9COJ
 
 | | SH3-like tandem domain of human KIN protein | | Descriptor: | DNA/RNA-binding protein KIN17 | | Authors: | de Lourenco, I.O, Fossey, M.A, Souza, F.P, Seixas, F.A.V, Fernandez, M.A, Almeida, F.C.L, Caruso, I.P. | | Deposit date: | 2024-07-16 | | Release date: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3-like tandem domain of human KIN protein and its interaction with RNA homo-oligonucleotides
To Be Published
|
|
6UEQ
 
 | | Structure of TBP bound to C-C mismatch containing TATA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*CP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), SULFATE ION, ... | | Authors: | Schumacher, M.A, Al-Hashimi, H. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
6UL8
 
 | | RIP2 kinase catalytic domain complex with (5S,6S,8R)-2-(benzo[d]thiazol-5-yl)-6-hydroxy-4,5,6,7,8,9-hexahydro-5,8-methanopyrazolo[1,5-a][1,3]diazocine-3-carboxamide | | Descriptor: | (5S,6S,8R)-2-(1,3-benzothiazol-5-yl)-6-hydroxy-4,5,6,7,8,9-hexahydro-5,8-methanopyrazolo[1,5-a][1,3]diazocine-3-carboxamide, CALCIUM ION, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Shewchuk, L.M, Convery, M.A. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Pyrazolocarboxamides as Potent and Selective Receptor Interacting Protein 2 (RIP2) Kinase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
8TV4
 
 | | NMR structure of temporin L in solution | | Descriptor: | Temporin-1Tl peptide | | Authors: | McShan, A.C, Jia, R, Halim, M.A. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiviral peptides inhibiting the main protease of SARS-CoV-2 investigated by computational screening and in vitro protease assay.
J.Pept.Sci., 30, 2024
|
|
5LUE
 
 | | Minor form of the recombinant cytotoxin-1 from N. oxiana | | Descriptor: | VC-1=CYTOTOXIN | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
7LEN
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFS
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region with A265V mutation in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Epidermal growth factor receptor, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFR
 
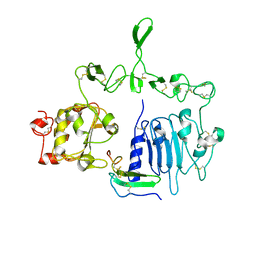 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with spermine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Proepiregulin, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
2Q0S
 
 | | Structure of the Inhibitor bound form of M. Smegmatis Aryl Esterase | | Descriptor: | Aryl esterase, SULFATE ION | | Authors: | Mathews, I.I, Soltis, M, Saldajeno, M, Ganshaw, G, Sala, R, Weyler, W, Cervin, M.A, Whited, G, Bott, R. | | Deposit date: | 2007-05-22 | | Release date: | 2007-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions.
Biochemistry, 46, 2007
|
|
2PUE
 
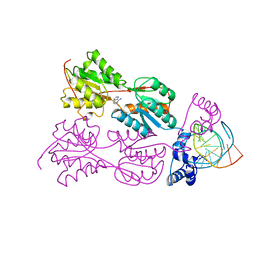 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | ADENINE, DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), PROTEIN (PURINE REPRESSOR ) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
7LQP
 
 | |
2Q0Q
 
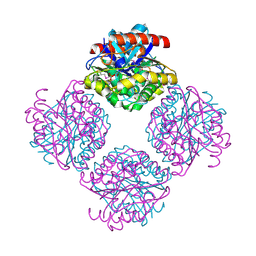 | | Structure of the Native M. Smegmatis Aryl Esterase | | Descriptor: | GLYCEROL, SULFATE ION, aryl esterase | | Authors: | Mathews, I.I, Soltis, M, Saldajeno, M, Ganshaw, G, Sala, R, Weyler, W, Cervin, M.A, Whited, G, Bott, R. | | Deposit date: | 2007-05-22 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions.
Biochemistry, 46, 2007
|
|
