3ZNQ
 
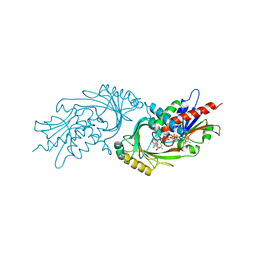 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 3-PHENETHYL-4H-FURO[3,2-B]PYRROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
6L3H
 
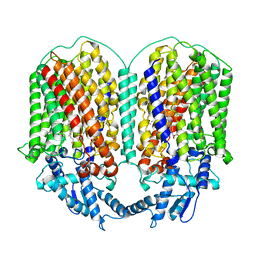 | | Cryo-EM structure of dimeric quinol dependent Nitric Oxide Reductase (qNOR) from the pathogen Neisseria meninigitidis | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Gopalasingam, C.C, Johnson, R.M, Tosha, T, Muench, S.P, Muramoto, K, Antonyuk, S.V, Shiro, Y, Hasnain, S.S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
6MEB
 
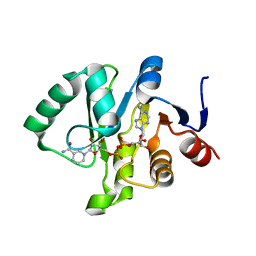 | | Crystal structure of Tylonycteris bat coronavirus HKU4 macrodomain in complex with nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
8ODU
 
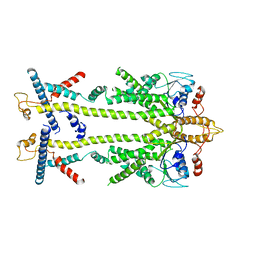 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (amphipol) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODV
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (nanodisc) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
4MF3
 
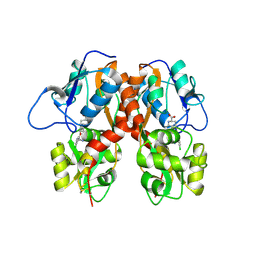 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4V3N
 
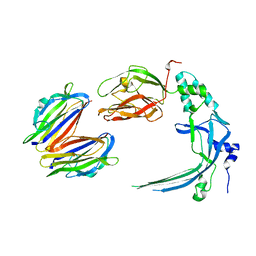 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V54
 
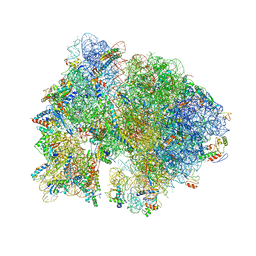 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
5IWH
 
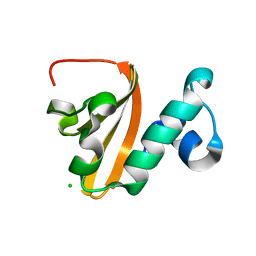 | | Structure of P. vulgaris HigB toxin delta H92 | | Descriptor: | CHLORIDE ION, Endoribonuclease HigB | | Authors: | Schureck, M.A, Repack, A.A, Miles, S.J, Marquez, J, Dunham, C.M. | | Deposit date: | 2016-03-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Mechanism of endonuclease cleavage by the HigB toxin.
Nucleic Acids Res., 44, 2016
|
|
5IXL
 
 | | Structure of P. vulgaris HigB toxin Y91A variant | | Descriptor: | CHLORIDE ION, Endoribonuclease HigB | | Authors: | Schureck, M.A, Repack, A.A, Miles, S.J, Marquez, J, Dunham, C.M. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of endonuclease cleavage by the HigB toxin.
Nucleic Acids Res., 44, 2016
|
|
5J79
 
 | | The identification and pharmacological characterization of 6-(tert-butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a highly potent and selective inhibitor of RIP2 Kinase, Compound 3 complex | | Descriptor: | 4-methyl-3-{[6-(methylsulfonyl)quinolin-4-yl]amino}phenol, Receptor-interacting serine/threonine-protein kinase 2, SULFATE ION | | Authors: | Convery, M.A, Casillas, L.N, Haile, P.A, Votta, B.J, Lakdawala, A.S. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The Identification and Pharmacological Characterization of 6-(tert-Butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a Highly Potent and Selective Inhibitor of RIP2 Kinase.
J.Med.Chem., 59, 2016
|
|
4U9G
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)CO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | CARBON MONOXIDE, NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9J
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NO-binding heme-dependent sensor protein, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9B
 
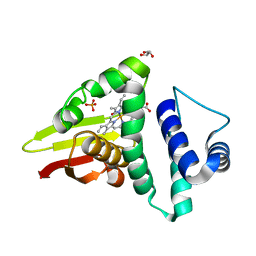 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9K
 
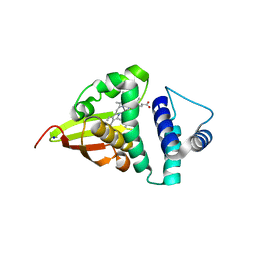 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II)NO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U99
 
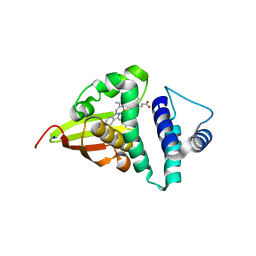 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IGH
 
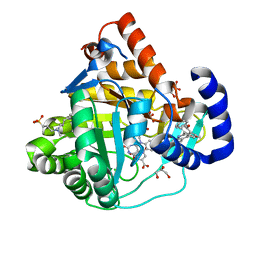 | | High resolution crystal structure of human dihydroorotate dehydrogenase bound with 4-quinoline carboxylic acid analog | | Descriptor: | 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, 6-fluoro-2-[2-methyl-4-phenoxy-5-(propan-2-yl)phenyl]quinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Das, P, Fontoura, B.M.A, Phillips, M.A, De Brabander, J.K. | | Deposit date: | 2012-12-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAR Based Optimization of a 4-Quinoline Carboxylic Acid Analog with Potent Anti-Viral Activity.
ACS MED.CHEM.LETT., 4, 2013
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
4XLL
 
 | |
7BJ4
 
 | | Inulosucrase from Halalkalicoccus jeotgali bound to kestose | | Descriptor: | Levansucrase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
7BJC
 
 | | Inulosucrase from Halalkalicoccus jeotgali in complex with sucrose | | Descriptor: | Levansucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
7BJ5
 
 | | Inulosucrase from Halalkalicoccus jeotgali | | Descriptor: | Levansucrase | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
7X9W
 
 | | Sulfur Oxygenase Reductase from Acidianus ambivalens | | Descriptor: | FE (III) ION, Sulfur oxygenase/reductase | | Authors: | Sobhy, M.A, Hamdan, S.M. | | Deposit date: | 2022-03-16 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Cryo-electron structures of the extreme thermostable enzymes Sulfur Oxygenase Reductase and Lumazine Synthase.
Plos One, 17, 2022
|
|
