1EMW
 
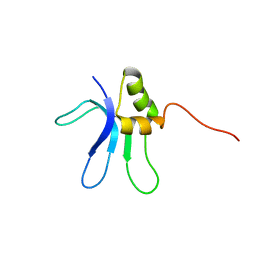 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S16 FROM THERMUS THERMOPHILUS | | Descriptor: | S16 RIBOSOMAL PROTEIN | | Authors: | Allard, P, Rak, A.V, Wimberly, B.T, Clemons Jr, W.M, Kalinin, A, Helgstrand, M, Garber, M.B, Ramakrishnan, V, Hard, T. | | Deposit date: | 2000-03-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Another piece of the ribosome: solution structure of S16 and its location in the 30S subunit.
Structure Fold.Des., 8, 2000
|
|
1MO9
 
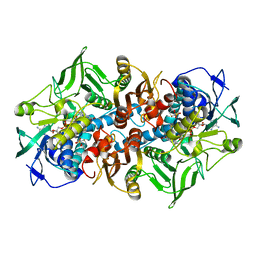 | | NADPH DEPENDENT 2-KETOPROPYL COENZYME M OXIDOREDUCTASE/CARBOXYLASE COMPLEXED WITH 2-KETOPROPYL COENZYME M | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, orf3 | | Authors: | Nocek, B, Jang, S.B, Jeong, M.S, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2002-09-08 | | Release date: | 2002-11-27 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for CO2 Fixation by a Novel Member of the Disulfide Oxidoreductase Family of Enzymes,
2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Biochemistry, 41, 2002
|
|
1MLX
 
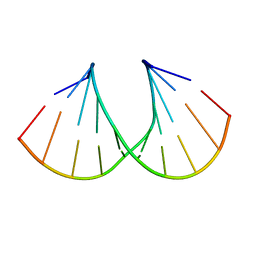 | | Crystal Structure Analysis of a 2'-O-[2-(Methylthio)-ethyl]-Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*SMTP*AP*CP*GP*C)-3' | | Authors: | Prakash, T.P, Manoharan, M, Kawasaki, A.M, Fraser, A.S, Lesnik, E.A, Sioufi, N, Leeds, J.M, Teplova, M, Egli, M. | | Deposit date: | 2002-08-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 2'-O-[2-(Methylthio)ethyl]-Modified Oligonucleotide: An Analogue of 2'-O-[2-(Methoxy)-ethyl]-Modified
Oligonucleotide with Improved Protein Binding Properties and High Binding Affinity to Target RNA
Biochemistry, 41, 2002
|
|
3VEB
 
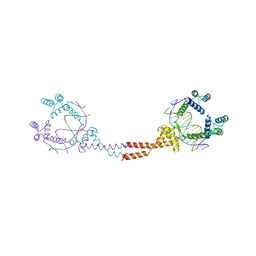 | | Crystal Structure of Matp-matS | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*G)-3', 5'-D(*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*G)-3', CALCIUM ION, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-01-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
1MKS
 
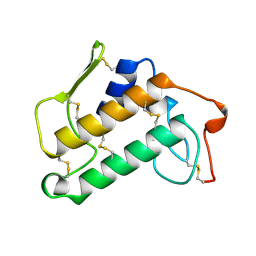 | |
1E56
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural substrate DIMBOA-beta-D-glucoside | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase- Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E66
 
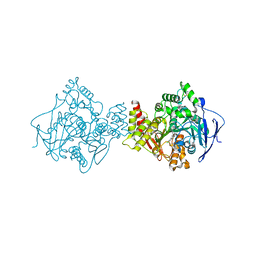 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT 2.1A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CHLORO-9-ETHYL-6,7,8,9,10,11-HEXAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-12-AMINE, ACETYLCHOLINESTERASE | | Authors: | Dvir, H, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D Structure of Torpedo Californica Acetylcholinesterase Complexed with Huprine X at 2. 1 A Resolution: Kinetic and Molecular Dynamic Correlates.
Biochemistry, 41, 2002
|
|
1E7Y
 
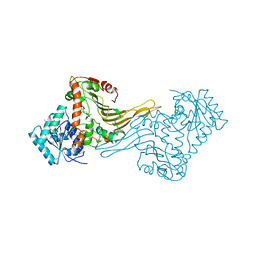 | | ACTIVE SITE MUTANT (D177->N) OF GLUCOSE 6-PHOSPHATE DEHYDROGENASE FROM LEUCONOSTOC MESENTEROIDES COMPLEXED WITH SUBSTRATE AND NADPH | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CALCIUM ION, GLUCOSE 6-PHOSPHATE 1-DEHYDROGENASE, ... | | Authors: | Adams, M.J, Cosgrove, M.S, Gover, S. | | Deposit date: | 2000-09-12 | | Release date: | 2000-12-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | An Examination of the Role of Asp-177 in the His-Asp Catalytic Dyad of Leuconostoc Mesenteroides Glucose 6-Phosphate Dehydrogenase: X-Ray Structure and Ph Dependence of Kinetic Parameters of the D177N Mutant Enzyme
Biochemistry, 39, 2000
|
|
1W9V
 
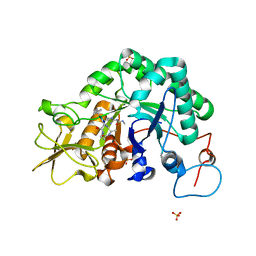 | | Specificity and affinity of natural product cyclopentapeptide argifin against Aspergillus fumigatus | | Descriptor: | ARGIFIN, CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1EL9
 
 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLTHIO]ACETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
5W24
 
 | | Crystal Structure of 5C4 Fab | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 5C4 Fab Heavy Chain, 5C4 Fab Light Chain | | Authors: | Battles, M.B, McLellan, J.S, Taher, N.M. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
1MU8
 
 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
1W9U
 
 | | Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against Aspergillus fumigatus chitinase | | Descriptor: | ARGADIN, CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
1EZ0
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
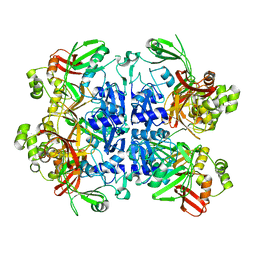
1EF8
 
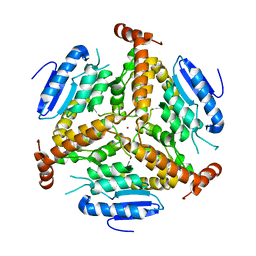 | | CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE | | Descriptor: | METHYLMALONYL COA DECARBOXYLASE, NICKEL (II) ION | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
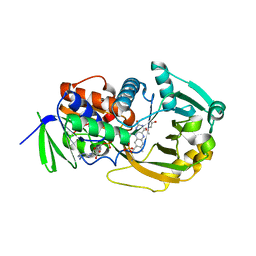
7N33
 
 | | SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state | | Descriptor: | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
1EL8
 
 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLSELENO]CETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
1EV1
 
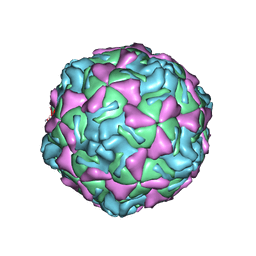 | | ECHOVIRUS 1 | | Descriptor: | ECHOVIRUS 1, MYRISTIC ACID, PALMITIC ACID | | Authors: | Wien, M.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-12-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure determination of echovirus 1.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1E55
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the competitive inhibitor dhurrin | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes.
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
5W8J
 
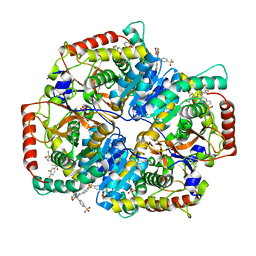 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 | | Descriptor: | 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lukacs, C.M, Moulin, A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
1W4N
 
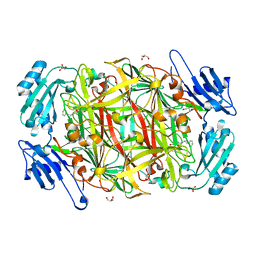 | | AGAO covalent complex with Tranylcypromine | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Duff, A.P, Trambaiolo, D.M, Langley, D.B, Juda, G.A, Shepard, E.M, Dooley, D.M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-07-27 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Complexes of the Copper-Containing Amine Oxidase from Arthrobacter Globiformis with the Inhibitors Benzylhydrazine and Tranylcypromine.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1VUB
 
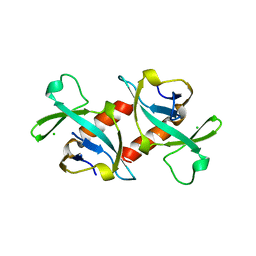 | | CCDB, A TOPOISOMERASE POISON FROM E. COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
3VFL
 
 | |
3DCQ
 
 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
