4UV2
 
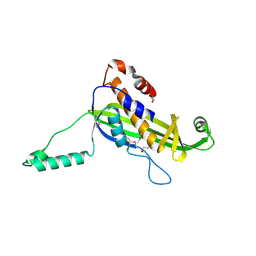 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
5JD2
 
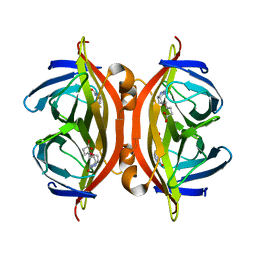 | | SFX structure of corestreptavidin-selenobiotin complex | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-selenopheno[3,4-d]imidazol-4-yl]pentanoic acid, Streptavidin | | Authors: | DeMirci, H, Hunter, M.S, Boutet, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser.
Nat Commun, 7, 2016
|
|
5JEG
 
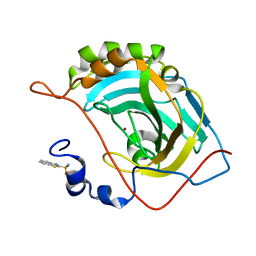 | | Human carbonic anhydrase II (V121I) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5IXQ
 
 | |
5JEP
 
 | | Human carbonic anhydrase II (T199S) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5IYX
 
 | |
5IZK
 
 | |
5JD3
 
 | | Crystal structure of LAE5, an alpha/beta hydrolase enzyme from the metagenome of Lake Arreo, Spain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, LAE5, ... | | Authors: | Stogios, P.J, Xu, X, Nocek, B, Cui, H, Yim, V, Martinez-Martinez, M, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
4UTB
 
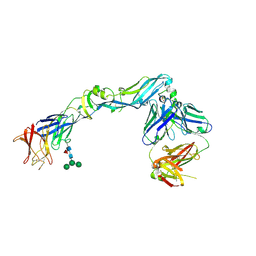 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UZ3
 
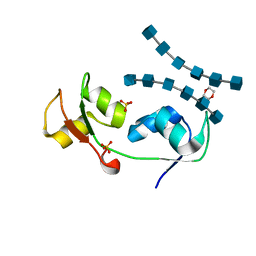 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Wong, J.E.M.M, Blaise, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UX9
 
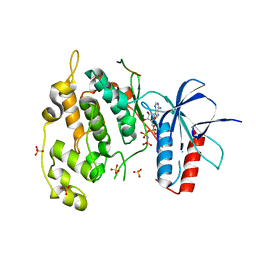 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5JED
 
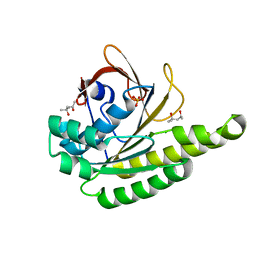 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
5JGD
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 12 | | Descriptor: | N-(2-isopropoxy-3-(4-methylpiperazine-1-carbonyl)phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
4UVX
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-fluoro-1,2-dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-fluoroisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVP
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- ethyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-ethylisoquinolin-1(2H)-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
5J4H
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
4UXZ
 
 | | Structure of delta7-DgkA-syn in 7.9 MAG to 2.18 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, ACETATE ION, ... | | Authors: | Li, D, Howe, N, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
4UZE
 
 | |
3KZL
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3L08
 
 | | Structure of Pi3K gamma with a potent inhibitor: GSK2126458 | | Descriptor: | 2,4-difluoro-N-[2-methoxy-5-(4-pyridazin-4-ylquinolin-6-yl)pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Marrero, E.M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
4UQH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
5J7Q
 
 | | Macrophage Migration Inhibitory Factor bound to Inhibitor K664 Derivative | | Descriptor: | 4-(imidazo[1,2-a]pyridin-2-yl)benzene-1,2-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Irregularities in enzyme assays: The case of macrophage migration inhibitory factor.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
