1MS4
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
3MCF
 
 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 3-alpha | | Descriptor: | CITRATE ANION, Diphosphoinositol polyphosphate phosphohydrolase 3-alpha, GLYCEROL | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-29 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 3-alpha
To be Published
|
|
1MTW
 
 | | FACTOR XA SPECIFIC INHIBITOR IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | (2S)-3-(7-carbamimidoylnaphthalen-2-yl)-2-[4-({(3R)-1-[(1Z)-ethanimidoyl]pyrrolidin-3-yl}oxy)phenyl]propanoic acid, CALCIUM ION, TRYPSIN | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of factor Xa specific inhibitors in complex with trypsin: structural grounds for inhibition of factor Xa and selectivity against thrombin.
FEBS Lett., 375, 1995
|
|
3VRK
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VS4
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
1MV1
 
 | | The Tandem, Sheared PA Pairs in 5'(rGGCPAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
2GEJ
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
1N1S
 
 | | Trypanosoma rangeli sialidase | | Descriptor: | SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-20 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T. cruzi
trans-sialidase
J.Mol.Biol., 325, 2003
|
|
6RV3
 
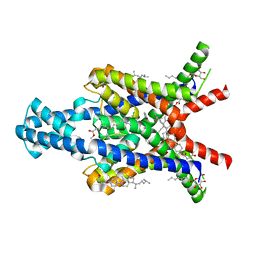 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6ELP
 
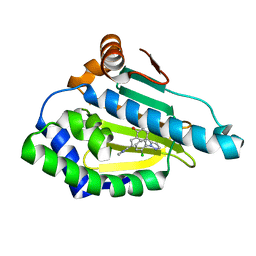 | |
5WO0
 
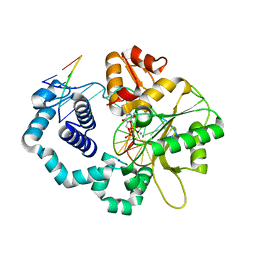 | | DNA polymerase beta substrate complex with incoming 5-FodUTP | | Descriptor: | 2'-deoxy-5-formyluridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
5A50
 
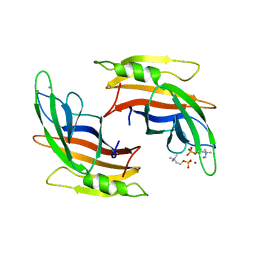 | |
2GEK
 
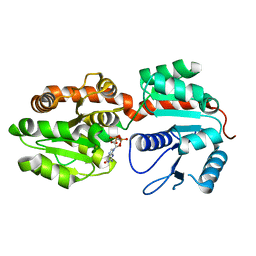 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
4WF6
 
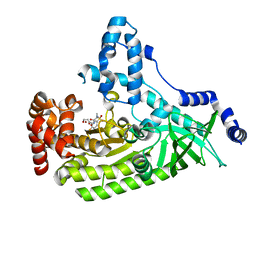 | | Anthrax toxin lethal factor with bound small molecule inhibitor MK-31 | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2014-09-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6521 Å) | | Cite: | Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates.
J.Med.Chem., 58, 2015
|
|
1MS9
 
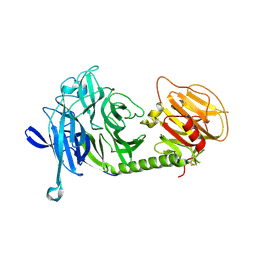 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme of Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MTS
 
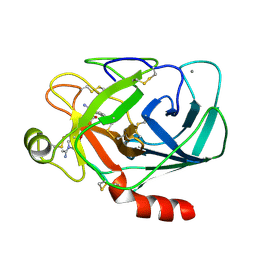 | | FACTOR XA SPECIFIC INHIBITOR IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | (+)-2-[4-[(-1-ACETIMIDOYL-4-PIPERIDINYL)OXY]-3-(7-AMIDINO-2-NAPHTHYL)PROPIONIC ACID, CALCIUM ION, TRYPSIN | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-16 | | Release date: | 1997-08-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of factor Xa specific inhibitors in complex with trypsin: structural grounds for inhibition of factor Xa and selectivity against thrombin.
FEBS Lett., 375, 1995
|
|
8HWI
 
 | | Bacterial STING from Larkinella arboricola in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
5I8F
 
 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
5AFE
 
 | |
6EL5
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
4PSB
 
 | |
2GK9
 
 | | Human Phosphatidylinositol-4-phosphate 5-kinase, type II, gamma | | Descriptor: | phosphatidylinositol-4-phosphate 5-kinase, type II, gamma | | Authors: | Uppenberg, J, Hogbom, M, Ogg, D, Arrowsmith, C, Berglund, H, Collins, R, Ehn, M, Flodin, S, Flores, A, Graslund, S, Holmberg-Schiavone, L, Edwards, A, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Human Phosphatidylinositol-4-phosphate 5-kinase, type II, gamma
To be Published
|
|
2GT7
 
 | | Crystal structure of SARS coronavirus main peptidase at pH 6.0 in the space group P21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
6RCJ
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OMe | | Descriptor: | GLYCEROL, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1E83
 
 | | Cytochrome c' from Alcaligenes xylosoxidans - oxidized structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
