7JJ8
 
 | | Crystal structure of the Zn(II)-bound ZnuA-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
4ZUZ
 
 | | SidC 1-871 | | Descriptor: | SidC | | Authors: | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
7T4Z
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in ligand-free form | | Descriptor: | AMMONIUM ION, GLYCEROL, Molybdate-binding periplasmic protein, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
4TRG
 
 | | the SNL domain of SidC | | Descriptor: | MERCURY (II) ION, SidC | | Authors: | Hsu, F.S, Luo, X, Qiu, J, Teng, Y, Jin, J, Smolka, M.B, Luo, Z.Q, Mao, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TRH
 
 | | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling | | Descriptor: | SidC | | Authors: | Hsu, F.S, Luo, X, Qiu, J, Teng, Y, Jin, J, Smolka, M.B, Luo, Z.Q, Mao, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7LVP
 
 | | Cryptococcus neoformans AIR synthetase | | Descriptor: | 1,2-ETHANEDIOL, AIR synthase, GLYCINE, ... | | Authors: | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
7LVO
 
 | | Cryptococcus neoformans GAR synthetase | | Descriptor: | 1,2-ETHANEDIOL, phosphoribosyl-glycinamide (GAR) synthetase | | Authors: | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
7LCY
 
 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | Descriptor: | Isoform B of NAD(+) hydrolase sarm1 | | Authors: | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
7T51
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in molybdate-bound form | | Descriptor: | AMMONIUM ION, GLYCEROL, MOLYBDATE ION, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-11 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T50
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in chromate-bound form | | Descriptor: | AMMONIUM ION, Chromate, GLYCEROL, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-11 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T5A
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in tungstate-bound form | | Descriptor: | AMMONIUM ION, Molybdate-binding periplasmic protein ModA, TUNGSTATE(VI)ION | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-11 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7LCZ
 
 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | Descriptor: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | Authors: | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7L5S
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7L5I
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7M6K
 
 | |
6UMP
 
 | | Crystal structure of MavC in complex with substrate mimic in P65 space group | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Luo, Z.Q, Das, C. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
6A6B
 
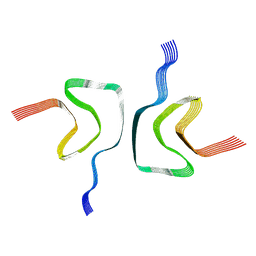 | | cryo-em structure of alpha-synuclein fiber | | Descriptor: | Alpha-synuclein | | Authors: | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
6KJ4
 
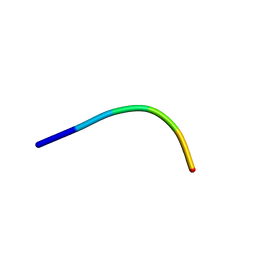 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ2
 
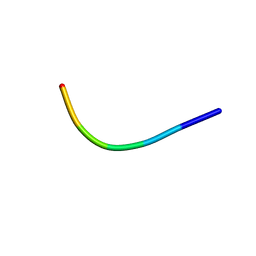 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ1
 
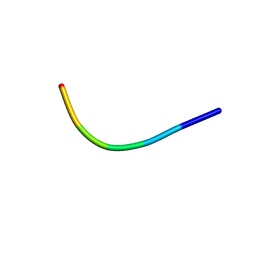 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
4FYG
 
 | | Structural basis for substrate recognition by a novel Legionella phosphoinositide phosphatase | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FYE
 
 | | Crystal structure of a Legionella phosphoinositide phosphatase, SidF | | Descriptor: | PHOSPHATE ION, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
