4UTO
 
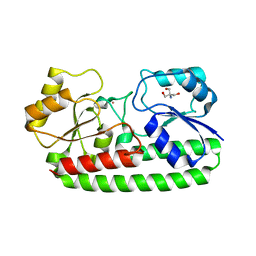 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4UTP
 
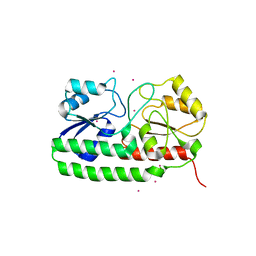 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
6LI2
 
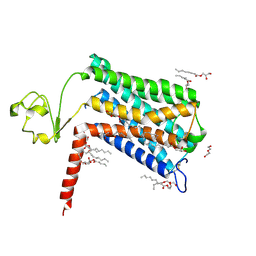 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
8STB
 
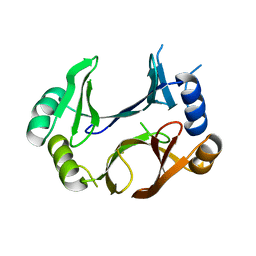 | | The structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | CHLORIDE ION, GLYCEROL, Glyoxalase, ... | | Authors: | Luo, Z, Jia, X, Yan, X, Qu, X, Kobe, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The crystal structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX.
To Be Published
|
|
6W0S
 
 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
7SHJ
 
 | |
6VZB
 
 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
5HN2
 
 | |
6VXV
 
 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
5HNJ
 
 | |
6VZA
 
 | | Crystal structure of cytochrome P450 NasF5053 Q65I-A86G mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
7C7M
 
 | | The structure of SAM-bound CntL, an aminobutyrate transferase in staphylopine biosysnthesis | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYLMETHIONINE, Staphylopine biosynthesis enzyme CntL | | Authors: | Luo, Z, Luo, S, Zhou, H. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis.
Faseb J., 35, 2021
|
|
8W9I
 
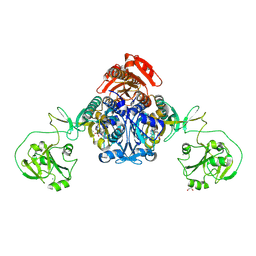 | |
8W8J
 
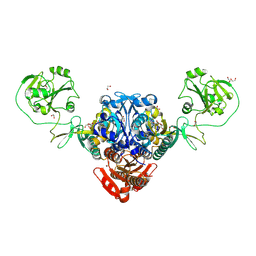 | |
6CSL
 
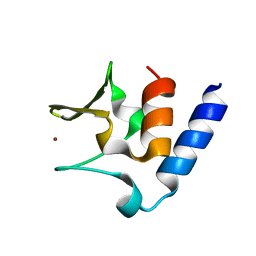 | | Pneumococcal PhtD protein 269-339 fragment with bound Zn(II) | | Descriptor: | Histidine triad protein D, ZINC ION | | Authors: | Luo, Z, Pederick, V.G, Paton, J.C, McDevitt, C.A, Kobe, B. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural characterisation of the HT3 motif of the polyhistidine triad protein D from Streptococcus pneumoniae.
FEBS Lett., 592, 2018
|
|
7C9M
 
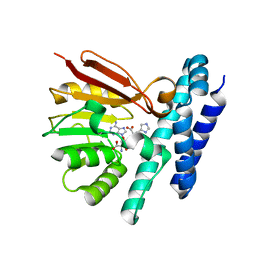 | | The structure of product-bound CntL, an aminobutyrate transferase in staphylopine biosynthesis | | Descriptor: | (2S)-2-azanyl-4-[[(2R)-3-(1H-imidazol-4-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]butanoic acid, 5'-DEOXY-5'-METHYLTHIOADENOSINE, D-histidine 2-aminobutanoyltransferase | | Authors: | Luo, Z, Zhou, H. | | Deposit date: | 2020-06-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis.
Faseb J., 35, 2021
|
|
7C9K
 
 | | Crystal Structure of E84Q mutant of CntL in complex with SAM | | Descriptor: | CALCIUM ION, D-histidine 2-aminobutanoyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Luo, Z, Zhou, H. | | Deposit date: | 2020-06-06 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis.
Faseb J., 35, 2021
|
|
8H7J
 
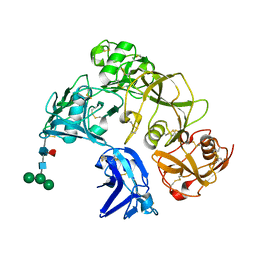 | | The crystal structure of CD163 SRCR5-9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor cysteine-rich type 1 protein M130, ... | | Authors: | Luo, Z.P, Li, R, Ma, H.F. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of CD163 SRCR5-9
To Be Published
|
|
5HNQ
 
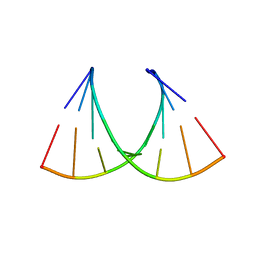 | |
8W8L
 
 | | Crystal structure of bacterial prolyl-tRNA synthetase in complex with inhibitor PAA-38 | | Descriptor: | (2~{S})-~{N}-[5-(4-azanyl-8-fluoranyl-quinazolin-7-yl)-2-fluoranyl-phenyl]sulfonylpyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Luo, Z, Zhou, H. | | Deposit date: | 2023-09-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structure-Guided Fluorine Scanning Accelerates the Discovery of Potent and Selective Inhibitors Against Bacterial Prolyl-tRNA Synthetase
To Be Published
|
|
8WWQ
 
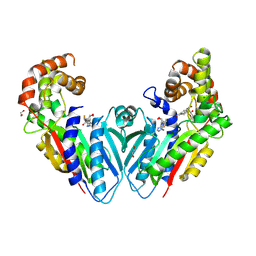 | | geniposidic acid O-methyltransferase complexed with SAH and geniposidic acid | | Descriptor: | 1,2-ETHANEDIOL, Geniposidic acid, MAGNESIUM ION, ... | | Authors: | Luo, Z, Li, L, Ma, D, Zhou, H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the substrate specificity of GAMT and divergence of seco-iridoid and closed-ring iridoid
To Be Published
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
7JJ8
 
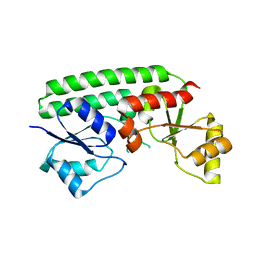 | | Crystal structure of the Zn(II)-bound ZnuA-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
6N8A
 
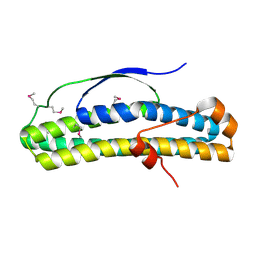 | | Crystal structure of selenomethionine-containing AcaB from uropathogenic E. coli | | Descriptor: | CHLORIDE ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-28 | | Release date: | 2020-07-15 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
