3HGW
 
 | | Apo Structure of Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase I87T mutant | | Descriptor: | CALCIUM ION, Salicylate biosynthesis protein pchB | | Authors: | Luo, Q, Lamb, A.L. | | Deposit date: | 2009-05-14 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-function analyses of isochorismate-pyruvate lyase from Pseudomonas aeruginosa suggest differing catalytic mechanisms for the two pericyclic reactions of this bifunctional enzyme
Biochemistry, 48, 2009
|
|
7BP4
 
 | |
7BP6
 
 | |
7BP2
 
 | |
7BP5
 
 | |
5X5Y
 
 | | A membrane protein complex | | Descriptor: | Probable ATP-binding component of ABC transporter, Uncharacterized protein | | Authors: | Luo, Q, Yang, X, Huang, Y. | | Deposit date: | 2017-02-18 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.465 Å) | | Cite: | Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG
Nat. Struct. Mol. Biol., 24, 2017
|
|
3HGX
 
 | |
8H1R
 
 | | Crystal structure of LptDE-YifL complex | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
8H1S
 
 | | Crystal structure of apo-LptDE complex | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
6M2M
 
 | |
7C7X
 
 | |
7CV9
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CVA
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV6
 
 | | RNA methyltransferase METTL4 | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV8
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, SINEFUNGIN | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV7
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7DPE
 
 | | RNA methyltransferase METTL4 | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*GP*TP*GP*AP*TP*CP*AP*CP*GP*CP*GP*GP*C)-3'), GLYCEROL, Methyltransferase-like protein 2, ... | | Authors: | Luo, Q, Ma, J.B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structure of RNA m6A methyltransferase METTL4 in complex with DNA at 2.75 Angstroms resolutioon
To Be Published
|
|
7EFO
 
 | | LptB2FG-LPS from Klebsiella pneumoniae in nanodiscs | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, LPS export ABC transporter permease LptG, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Luo, Q, Shi, H. | | Deposit date: | 2021-03-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structures of LptB 2 FG and LptB 2 FGC from Klebsiella pneumoniae in complex with lipopolysaccharide.
Biochem.Biophys.Res.Commun., 571, 2021
|
|
7FDN
 
 | |
4HTM
 
 | | Mechanism of CREB Recognition and Coactivation by the CREB Regulated Transcriptional Coactivator CRTC2 | | Descriptor: | CREB-regulated transcription coactivator 2, ZINC ION | | Authors: | Luo, Q, Viste, K, Urday-Zaa, J.C, Kumar, G.S, Tsai, W.-W, Talai, A, Mayo, K, Montminy, M, Radhakrishnan, I. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein kinase N1, a cell inhibitor of Akt kinase, has a central role in quality control of germinal center formation
Proc.Natl.Acad.Sci.USA, 2012
|
|
7FDM
 
 | |
7FDL
 
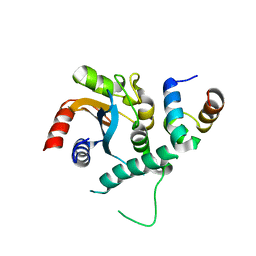 | |
8BNZ
 
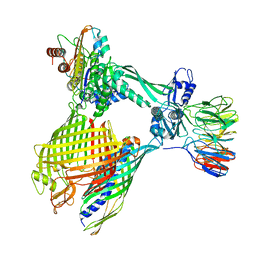 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
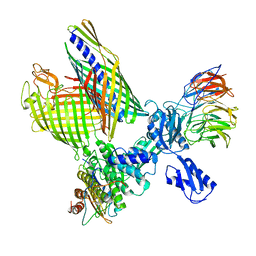 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
6KKS
 
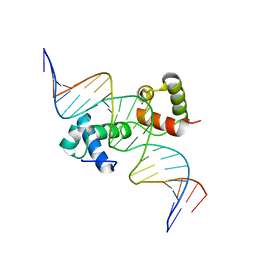 | | Structural insights into target DNA recognition by R2R3-type MYB transcription factor | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*CP*TP*CP*CP*AP*AP*CP*CP*GP*CP*AP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*CP*GP*AP*AP*AP*AP*TP*GP*CP*GP*GP*TP*TP*GP*GP*AP*GP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Wang, B, Luo, Q. | | Deposit date: | 2019-07-27 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into target DNA recognition by R2R3-MYB transcription factors.
Nucleic Acids Res., 48, 2020
|
|
