2R8T
 
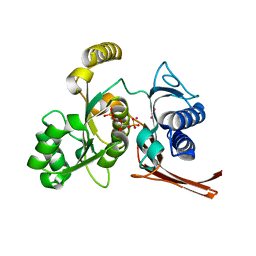 | | Crystal structure of the fructose 1,6-bisphosphatase GlpX from E.coli in the complex with fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase class II glpX, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-09-11 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fructose 1,6-bisphosphatase GlpX
from E.coli in the complex with fructose 1,6-bisphosphate
To be Published
|
|
2QGA
 
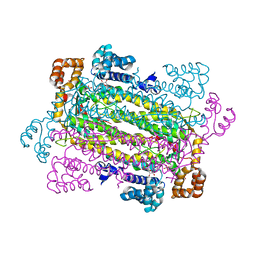 | | Plasmodium vivax adenylosuccinate lyase Pv003765 with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, CALCIUM ION, ... | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Hills, T, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Plasmodium vivax adenylosuccinate lyase Pv003765 with AMP bound
To be Published
|
|
1RWF
 
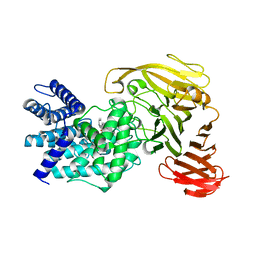 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWG
 
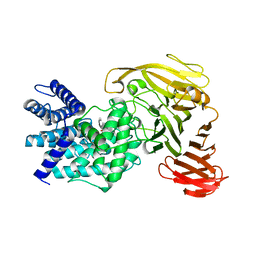 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1Q18
 
 | | Crystal structure of E.coli glucokinase (Glk) | | Descriptor: | Glucokinase | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.
J.Bacteriol., 186, 2004
|
|
1RW9
 
 | | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | PHOSPHATE ION, SODIUM ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Linhardt, R.J, Miyazono, H, Kyogashima, M, Kaneko, T, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1SKO
 
 | | MP1-p14 Complex | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Lunin, V.V, Munger, C, Wagner, J, Ye, Z, Cygler, M, Sacher, M. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the MAP kinase scaffold MP1 bound to its partner p14: a complex with a critical role in endosomal MAP kinase signaling
J.Biol.Chem., 279, 2004
|
|
1RWH
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWA
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | GLYCEROL, MERCURY (II) ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWC
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1SZ2
 
 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
2ESN
 
 | | The crystal structure of probable transcriptional regulator PA0477 from Pseudomonas aeruginosa | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Chang, C, Skarina, T, Gorodischenskaya, E, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of putative transcriptional regulator Pa0477 from Pseudomonas aeruginosa
To be Published
|
|
2FA1
 
 | | Crystal structure of PhnF C-terminal domain | | Descriptor: | Probable transcriptional regulator phnF, beta-D-fructopyranose | | Authors: | Lunin, V.V, Nocek, B.P, Gorelik, M, Skarina, T, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of GntR/HutC family signaling domain.
Protein Sci., 15, 2006
|
|
2FBH
 
 | | The crystal structure of transcriptional regulator PA3341 | | Descriptor: | MERCURY (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator PA3341
To be Published
|
|
2G7S
 
 | | The crystal structure of transcriptional regulator, TetR family, from Agrobacterium tumefaciens | | Descriptor: | transcriptional regulator, TetR family | | Authors: | Lunin, V.V, Chang, C, Xu, X, Gu, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of transcriptional regulator, TetR family, from Agrobacterium tumefaciens
To be Published
|
|
2FBQ
 
 | | The crystal structure of transcriptional regulator PA3006 | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Skarina, T, Onopriyenko, O, Kim, Y, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator PA3006
To be Published
|
|
2FBI
 
 | | The crystal structure of transcriptional regulator PA4135 | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Cuff, M.E, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of transcriptional regulator PA4135
To be Published
|
|
2FBL
 
 | | The crystal structure of the hypothetical protein NE1496 | | Descriptor: | SODIUM ION, hypothetical protein NE1496 | | Authors: | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the hypothetical protein NE1496
To be Published
|
|
1Y6Z
 
 | | Middle domain of Plasmodium falciparum putative heat shock protein PF14_0417 | | Descriptor: | heat shock protein, putative | | Authors: | Lunin, V.V, Botchkareva, E, Loppnau, P, Amani, M, Bray, J, Vedadi, M, Edwards, A, Arrowsmith, C, Sundstrom, M, Bochkarev, A, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-07 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1KK9
 
 | | CRYSTAL STRUCTURE OF E. COLI YCIO | | Descriptor: | SULFATE ION, probable translation factor yciO | | Authors: | Jia, J, Lunin, V.V, Sauve, V, Huang, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-06 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the YciO protein from Escherichia coli
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
5BV9
 
 | |
4WA0
 
 | |
6VSP
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A | | Descriptor: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
5TMA
 
 | | Zymomonas mobilis pyruvate decarboxylase mutant PDC-2.3 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An iterative computational design approach to increase the thermal endurance of a mesophilic enzyme.
Biotechnol Biofuels, 11, 2018
|
|
6XEW
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase | | Descriptor: | 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-06-14 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
