7OGL
 
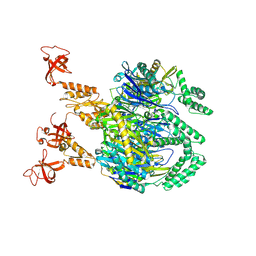 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. apo-PNPase | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7OGK
 
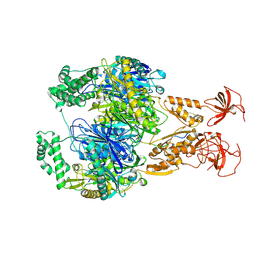 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ) | | Descriptor: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
286D
 
 | |
297D
 
 | |
2AAO
 
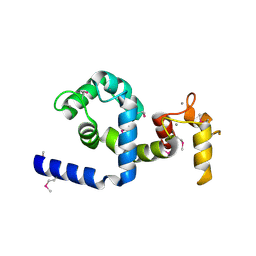 | | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, isoform AK1 | | Authors: | Chandran, V, Stollar, E.J, Lindorff-Larsen, K, Harper, J.F, Chazin, W.J, Dobson, C.M, Luisi, B.F, Christodoulou, J. | | Deposit date: | 2005-07-13 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the regulatory apparatus of a calcium-dependent protein kinase (CDPK): a novel mode of calmodulin-target recognition.
J.Mol.Biol., 357, 2006
|
|
285D
 
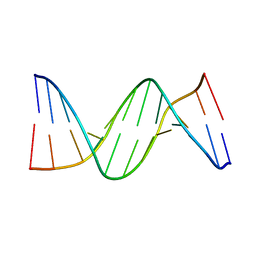 | |
4RMO
 
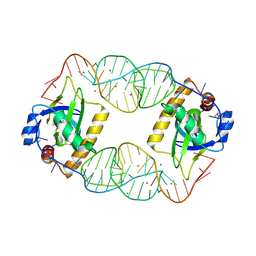 | | Crystal Structure of the CptIN Type III Toxin-Antitoxin System from Eubacterium rectale | | Descriptor: | CALCIUM ION, CptN Toxin, RNA (45-MER) | | Authors: | Rao, F, Voss, J.E, Short, F.L, Luisi, B.F. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-30 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system.
Nucleic Acids Res., 43, 2015
|
|
5V5S
 
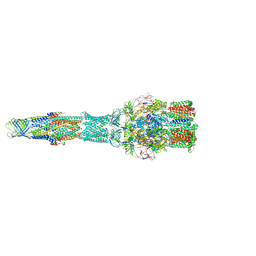 | | multi-drug efflux; membrane transport; RND superfamily; Drug resistance | | Descriptor: | Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, Outer membrane protein TolC | | Authors: | wang, Z, fan, G, Hryc, C.F, Blaza, J.N, Serysheva, I.I, Schmid, M.F, Chiu, W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-15 | | Release date: | 2017-04-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC Multidrug Efflux Pump.
Elife, 6, 2017
|
|
1P7J
 
 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1W88
 
 | | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, PYRUVATE DEHYDROGENASE E1 COMPONENT, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Switch and Proton-Wire Synchronize the Active Sites in Thiamine-Dependent Enzymes
Science, 306, 2004
|
|
1W85
 
 | | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
Science, 306, 2004
|
|
6O1L
 
 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression Hfq-Crc-amiE 2:3:2 complex | | Descriptor: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | Authors: | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | Deposit date: | 2019-02-20 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
6O1K
 
 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:2:2 complex (core complex) | | Descriptor: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | Authors: | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | Deposit date: | 2019-02-20 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Architectural principles for Hfq/Crc-mediated regulation of gene expression
Elife, 8, 2019
|
|
6O1M
 
 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:4:2 complex | | Descriptor: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | Authors: | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | Deposit date: | 2019-02-20 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
3H8A
 
 | |
4O7J
 
 | |
4QKK
 
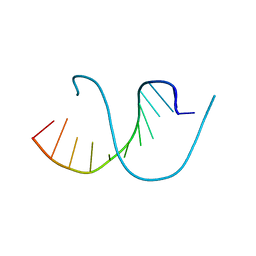 | | Crystal structure of an oligonucleotide containing 5-formylcytosine | | Descriptor: | DNA (5'-D(*CP*TP*AP*(5FC)P*GP*(5FC)P*GP*(5FC)P*GP*TP*AP*G)-3') | | Authors: | Raiber, E.-A, Murat, P, Chirgadze, D.Y, Luisi, B.F, Balasubramanian, S. | | Deposit date: | 2014-06-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 5-Formylcytosine alters the structure of the DNA double helix.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3DVA
 
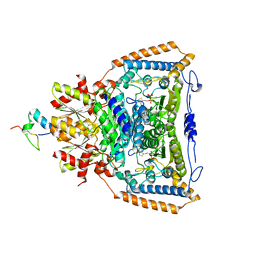 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3CFS
 
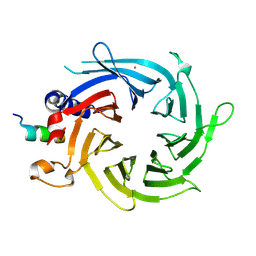 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | Authors: | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
3CFV
 
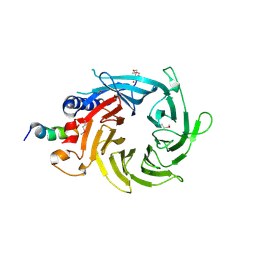 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4 peptide, Histone-binding protein RBBP7 | | Authors: | Pei, X.-Y, Murzina, N.V, Zhang, W, McLaughlin, S, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
3DUF
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DV0
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
2FYM
 
 | |
1TRO
 
 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
