6QSU
 
 | | Helicobacter pylori urease with BME bound in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2019-02-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
6ZJA
 
 | | Helicobacter pylori urease with inhibitor bound in the active site | | Descriptor: | 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-12-23 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
1C8S
 
 | | BACTERIORHODOPSIN D96N LATE M STATE INTERMEDIATE | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN ("M" STATE INTERMEDIATE), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
1C8R
 
 | | BACTERIORHODOPSIN D96N BR STATE AT 2.0 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, PROTEIN (BACTERIORHODOPSIN), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
1C3W
 
 | | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.55 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN (GROUND STATE WILD TYPE "BR"), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-28 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of bacteriorhodopsin at 1.55 A resolution.
J.Mol.Biol., 291, 1999
|
|
1F50
 
 | | BACTERIORHODOPSIN-BR STATE OF THE E204Q MUTANT AT 1.7 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Luecke, H, Schobert, B, Cartailler, J.P, Richter, H.T, Rosengarth, A, Needleman, R, Lanyi, J.K. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin.
J.Mol.Biol., 300, 2000
|
|
1F4Z
 
 | | BACTERIORHODOPSIN-M PHOTOINTERMEDIATE STATE OF THE E204Q MUTANT AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Luecke, H, Schobert, B, Cartailler, J.P, Richter, H.T, Rosengarth, A, Needleman, R, Lanyi, J.K. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin.
J.Mol.Biol., 300, 2000
|
|
2NOM
 
 | |
1MCX
 
 | |
1JGJ
 
 | |
1AEI
 
 | | CRYSTAL STRUCTURE OF THE ANNEXIN XII HEXAMER | | Descriptor: | ANNEXIN XII, CALCIUM ION | | Authors: | Luecke, H, Chang, B.T, Mailliard, W.S, Schlaepfer, D.D, Haigler, H.T. | | Deposit date: | 1995-09-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the annexin XII hexamer and implications for bilayer insertion.
Nature, 378, 1995
|
|
1BRX
 
 | | BACTERIORHODOPSIN/LIPID COMPLEX | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Luecke, H, Richter, H.T, Lanyi, J. | | Deposit date: | 1998-05-28 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution.
Science, 280, 1998
|
|
2IKF
 
 | |
1HM6
 
 | |
7Q4Q
 
 | | Magacizumab Fab fragment in complex with human LRG1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRG1 epitope, Magacizumab heavy chain, ... | | Authors: | Gutierrez-Fernandez, J, Luecke, H. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of human LRG1 recognition by Magacizumab, a humanized monoclonal antibody with therapeutic potential.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4TL3
 
 | |
5KAL
 
 | | Terminal uridylyl transferase 4 from Trypanosoma brucei with bound UTP and UpU | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*U)-3'), RNA uridylyltransferase 4, ... | | Authors: | Stagno, J.R, Luecke, H, Afasizhev, R. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
3P7N
 
 | |
2Q0E
 
 | |
2Q0D
 
 | |
2Q0G
 
 | |
4FTG
 
 | |
1TP7
 
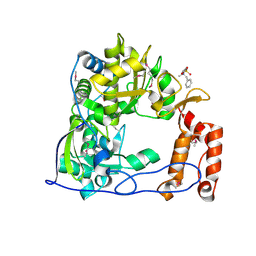 | | Crystal Structure of the RNA-dependent RNA Polymerase from Human Rhinovirus 16 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Genome polyprotein, SULFATE ION | | Authors: | Appleby, T.C, Luecke, H, Shim, J.H, Wu, J.Z, Cheney, I.W, Zhong, W, Vogeley, L, Hong, Z, Yao, N. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer.
J.Virol., 79, 2005
|
|
3T45
 
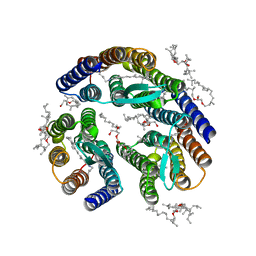 | | Crystal structure of bacteriorhodopsin mutant A215T, a phototaxis signaling mutant at 3.0 A resolution | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin (GROUND STATE), RETINAL | | Authors: | Ozorowski, G, Luecke, H. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A transporter converted into a sensor, a phototaxis signaling mutant of bacteriorhodopsin at 3.0 angstrom.
J.Mol.Biol., 415, 2012
|
|
3UX4
 
 | |
