1CV4
 
 | | T4 LYSOZYME MUTANT L118M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CVK
 
 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
8DU2
 
 | | HnRNPA2 D290V LCD PM1 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
1EH5
 
 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 COMPLEXED WITH PALMITATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8DUW
 
 | | HnRNPA2 D290V LCD PM2 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
3OMY
 
 | | Crystal structure of the pED208 TraM N-terminal domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein traM | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
8EC7
 
 | | HnRNPA2 D290V LCD PM3 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
3ON0
 
 | | Crystal structure of the pED208 TraM-sbmA complex | | Descriptor: | Protein traM, sbmA | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
8GZI
 
 | |
4NK3
 
 | | Amp-c beta-lactamase (pseudomonas aeruginosa) in complex with mk-7655 | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin().
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1CTW
 
 | | T4 LYSOZYME MUTANT I78A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV6
 
 | | T4 LYSOZYME MUTANT V149M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
7SUO
 
 | |
7SMV
 
 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
7SNA
 
 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
8GX4
 
 | |
3PLA
 
 | | Crystal structure of a catalytically active substrate-bound box C/D RNP from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L7Ae, C/D guide RNA, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, ... | | Authors: | Lin, J, Lai, S, Jia, R, Xu, A, Zhang, L, Lu, J, Ye, K. | | Deposit date: | 2010-11-15 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for site-specific ribose methylation by box C/D RNA protein complexes.
Nature, 469, 2011
|
|
1EI9
 
 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOYL PROTEIN THIOESTERASE 1 | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-24 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3K6J
 
 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
3S22
 
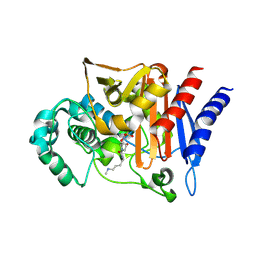 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with an inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, [(2S,3R)-2-formyl-1-{[4-(methylamino)butyl]carbamoyl}pyrrolidin-3-yl]sulfamic acid | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2LAS
 
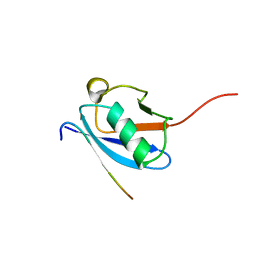 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
3S1Y
 
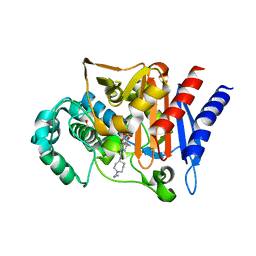 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with a beta-lactamase inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4QPO
 
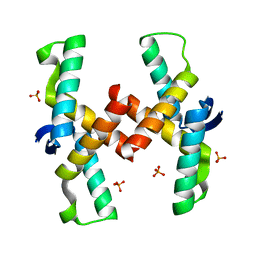 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | PHOSPHATE ION, Relaxosome protein TraM | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
1CU0
 
 | | T4 LYSOZYME MUTANT I78M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
3R7G
 
 | | Crystal structure of Spire KIND domain in complex with the tail of FMN2 | | Descriptor: | Formin-2, Protein spire homolog 1 | | Authors: | Kreutz, B, Vizcarra, C.L, Rodal, A.A, Toms, A.V, Lu, J, Quinlan, M.E, Eck, M.J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Spire KIND domain and insights into its interaction with Fmn-family formins
Proc.Natl.Acad.Sci.USA, 2011
|
|
