1ZJI
 
 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
3BE9
 
 | | Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities | | Descriptor: | 19-(cyclopropylamino)-4,6,7,15-tetrahydro-5H-16,1-(azenometheno)-10,14-(metheno)pyrazolo[4,3-o][1,3,9]triazacyclohexadecin-8(9H)-one, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Lu, J, Averill, A, Almassy, R, Chu, S. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3IL5
 
 | | Structure of E. faecalis FabH in complex with 2-({4-bromo-3-[(diethylamino)sulfonyl]benzoyl}amino)benzoic acid | | Descriptor: | 2-({[4-bromo-3-(diethylsulfamoyl)phenyl]carbonyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL6
 
 | | Structure of E. faecalis FabH in complex with 2-({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxybenzoyl}amino)benzoic acid | | Descriptor: | 2-[({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxyphenyl}carbonyl)amino]benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SULFATE ION | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL9
 
 | | Structure of E. coli FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
8T1U
 
 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
8T12
 
 | |
8T13
 
 | |
3IL7
 
 | | Crystal structure of S. aureus FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL3
 
 | | Structure of Haemophilus influenzae FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL4
 
 | | Structure of E. faecalis FabH in complex with acetyl CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, ACETYL COENZYME *A | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
7LCO
 
 | | Improved Feline Drugs as SARS-CoV-2 Mpro Inhibitors: Structure-Activity Studies & Micellar Solubilization for Enhanced Bioavailability | | Descriptor: | (3-fluorophenyl)methyl [(2S)-3-cyclopropyl-1-oxo-1-({(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Khan, M.B, Lu, J, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improved SARS-CoV-2 M pro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies.
Eur.J.Med.Chem., 222, 2021
|
|
7LDL
 
 | | Improved Feline Drugs as SARS-CoV-2 Mpro Inhibitors: Structure-Activity Studies & Micellar Solubilization for Enhanced Bioavailability | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improved SARS-CoV-2 M pro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies.
Eur.J.Med.Chem., 222, 2021
|
|
8DKZ
 
 | |
8DJJ
 
 | |
8DK8
 
 | |
1U8T
 
 | | Crystal structure of CheY D13K Y106W alone and in complex with a FliM peptide | | Descriptor: | Chemotaxis protein cheY, Flagellar motor switch protein fliM, SULFATE ION | | Authors: | Dyer, C.M, Quillin, M.L, Campos, A, Lu, J, McEvoy, M.M, Hausrath, A.C, Westbrook, E.M, Matsumura, P, Matthews, B.W, Dahlquist, F.W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Constitutively Active Double Mutant CheY(D13K Y106W) Alone and in Complex with a FliM Peptide
J.Mol.Biol., 342, 2004
|
|
3IU7
 
 | |
8GJN
 
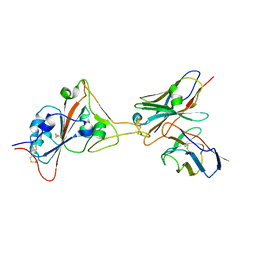 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
3IU9
 
 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T07 | | Descriptor: | 5-[(2,4-dichlorobenzyl)sulfanyl]-4H-1,2,4-triazol-3-amine, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
3IU8
 
 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T03 | | Descriptor: | 3-[(4-fluorobenzyl)sulfanyl]-4H-1,2,4-triazole, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
8FTC
 
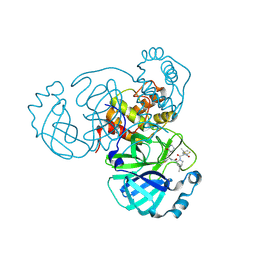 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | (1R,2S,5S)-3-[N-(difluoroacetyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopiperidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Chen, P, Khan, M.B, Lu, J, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Effect of Deuteration and Homologation of the Lactam Ring of Nirmatrelvir on Its Biochemical Properties and Oxidative Metabolism.
Acs Bio Med Chem Au, 3, 2023
|
|
8FZB
 
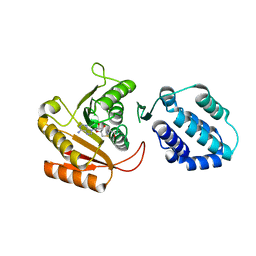 | | Crystal structure of human FAM86A | | Descriptor: | Protein-lysine N-methyltransferase EEF2KMT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shao, Z, Lu, J, Song, J. | | Deposit date: | 2023-01-28 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The FAM86 domain of FAM86A confers substrate specificity to promote EEF2-Lys525 methylation.
J.Biol.Chem., 299, 2023
|
|
2H9D
 
 | |
2H9C
 
 | |
