6TAD
 
 | | Bd0314 DslA E143Q mutant | | Descriptor: | SLT domain-containing protein | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
6TAF
 
 | | Bd0314 DslA E154Q mutant | | Descriptor: | ACETATE ION, SLT domain-containing protein, SULFATE ION | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
6SCI
 
 | | Structure of AdhE form 1 | | Descriptor: | Aldehyde-alcohol dehydrogenase, FE (III) ION | | Authors: | Lovering, A.L, Bragginton, E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TA9
 
 | | Bd0314 DslA wild-type form 1 | | Descriptor: | SLT domain-containing protein, SULFATE ION | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
1XSI
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETIC ACID, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XSK
 
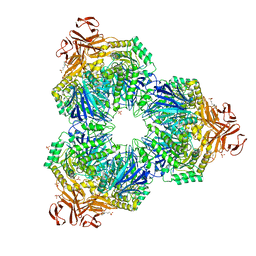 | | Structure of a Family 31 alpha glycosidase glycosyl-enzyme intermediate | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 5(R)-fluoro-beta-D-xylopyranose, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 {alpha}-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XSJ
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative family 31 glucosidase yicI | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1S1P
 
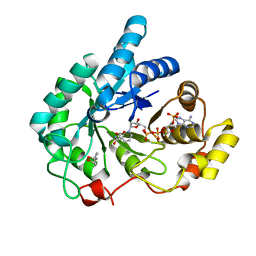 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1S1R
 
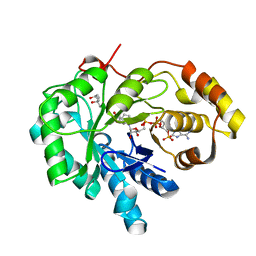 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1S2A
 
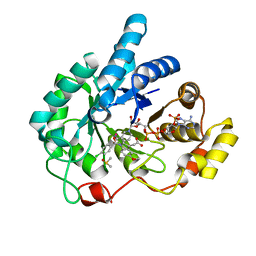 | | Crystal structures of prostaglandin D2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, INDOMETHACIN, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1S2C
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
4NX8
 
 | | Structure of a PTP-like phytase from Bdellovibrio bacteriovorus | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein-tyrosine phosphatase 2 | | Authors: | Gruninger, R.J, Lovering, A.L. | | Deposit date: | 2013-12-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural and biochemical analysis of a unique phosphatase from Bdellovibrio bacteriovorus reveals its structural and functional relationship with the protein tyrosine phosphatase class of phytase.
Plos One, 9, 2014
|
|
2F2H
 
 | | Structure of the YicI thiosugar Michaelis complex | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-NITROPHENYL 6-THIO-6-S-ALPHA-D-XYLOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, GLYCEROL, ... | | Authors: | Kim, Y.-W, Lovering, A.L, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2005-11-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expanding the Thioglycoligase Strategy to the Synthesis of alpha-linked Thioglycosides Allows Structural Investigation of the Parent Enzyme/Substrate Complex
J.Am.Chem.Soc., 128, 2006
|
|
2N5G
 
 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
6GFV
 
 | | M tuberculosis LpqI | | Descriptor: | Probable conserved lipoprotein LpqI | | Authors: | Moynihan, P.J, Lovering, A.L. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hydrolase LpqI primes mycobacterial peptidoglycan recycling.
Nat Commun, 10, 2019
|
|
6GKI
 
 | | Structure of E coli MlaC in Variously Loaded States | | Descriptor: | BROMIDE ION, GLYCEROL, Probable phospholipid-binding protein MlaC | | Authors: | Knowles, T.J, Lovering, A.L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system.
Nat Microbiol, 4, 2019
|
|
3L7J
 
 | |
3L7K
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (15 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Strynadka, N.C.J, Lovering, A.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L7I
 
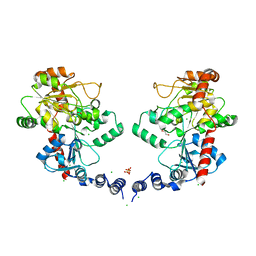 | |
7O0A
 
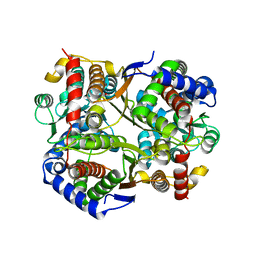 | | Bdellovibrio bacteriovorus PGI in P1211 spacegroup | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glucose-6-phosphate isomerase | | Authors: | Meek, R.W, Lovering, A.L. | | Deposit date: | 2021-03-26 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Bdellovibrio bacteriovorus phosphoglucose isomerase structures reveal novel rigidity in the active site of a selected subset of enzymes upon substrate binding.
Open Biology, 11, 2021
|
|
7NTG
 
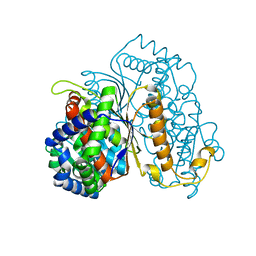 | | Bdellovibrio bacteriovorus PGI in complex with fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, FRUCTOSE -6-PHOSPHATE, Glucose-6-phosphate isomerase | | Authors: | Meek, R.W, Lovering, A.L. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Bdellovibrio bacteriovorus phosphoglucose isomerase structures reveal novel rigidity in the active site of a selected subset of enzymes upon substrate binding.
Open Biology, 11, 2021
|
|
7NSS
 
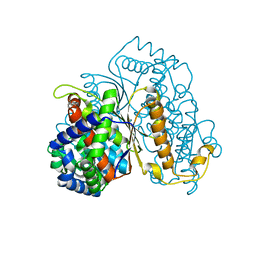 | | Bdellovibrio bacteriovorus PGI in P3121 spacegroup | | Descriptor: | 1,2-ETHANEDIOL, Glucose-6-phosphate isomerase | | Authors: | Meek, R.W, Lovering, A.L. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Bdellovibrio bacteriovorus phosphoglucose isomerase structures reveal novel rigidity in the active site of a selected subset of enzymes upon substrate binding.
Open Biology, 11, 2021
|
|
1YLU
 
 | | The structure of E. coli nitroreductase with bound acetate, crystal form 2 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YKI
 
 | | The structure of E. coli nitroreductase bound with the antibiotic nitrofurazone | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YLR
 
 | | The structure of E.coli nitroreductase with bound acetate, crystal form 1 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
