7MQP
 
 | | E. coli dihydrofolate reductase complexed with 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-4'-methyl[1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
5ECC
 
 | | Klebsiella pneumoniae DfrA1 complexed with NADPH and 6-ethyl-5-(3-(2-methoxy-5-(pyridin-4-yl)phenyl)prop-1-yn-1-yl)pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[2-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, CALCIUM ION, Dehydrofolate reductase type I, ... | | Authors: | Lombardo, M.N, Anderson, A.C. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structures of Trimethoprim-Resistant DfrA1 Rationalize Potent Inhibition by Propargyl-Linked Antifolates.
ACS Infect Dis, 2, 2016
|
|
7REB
 
 | | E. coli dihydrofolate reductase complexed with 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RGK
 
 | | DfrA5 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase type 5, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RGO
 
 | | DfrA5 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase type 5, GLYCEROL, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RGJ
 
 | | DfrA1 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, 5-[(3S)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, CALCIUM ION, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7REG
 
 | | DfrA1 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, CALCIUM ION, Dihydrofolate reductase type 1, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
5ECX
 
 | | Klebsiella pneumoniae DfrA1 complexed with NADPH and 6-ethyl-5-(3-(6-(pyridin-4-yl)benzo[d][1,3]dioxol-4-yl)but-1-yn-1-yl)pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-[(3~{S})-3-(6-pyridin-4-yl-1,3-benzodioxol-4-yl)but-1-ynyl]pyrimidine-2,4-diamine, Dehydrofolate reductase type I, GLYCEROL, ... | | Authors: | Lombardo, M.N, Anderson, A.C. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Trimethoprim-Resistant DfrA1 Rationalize Potent Inhibition by Propargyl-Linked Antifolates.
ACS Infect Dis, 2, 2016
|
|
4K22
 
 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
6QYH
 
 | | Structure of Apo HPAB from E.coli | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, CALCIUM ION, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
6QYI
 
 | | Structure of HPAB from E.coli in complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
6H6P
 
 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6H6O
 
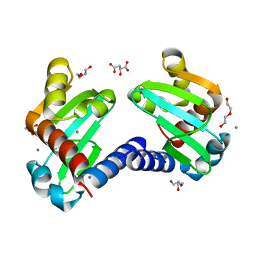 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6H6N
 
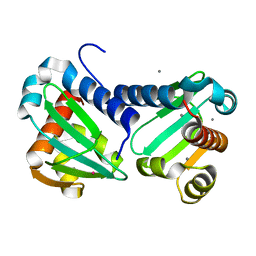 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, TERBIUM(III) ION, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
3ZQM
 
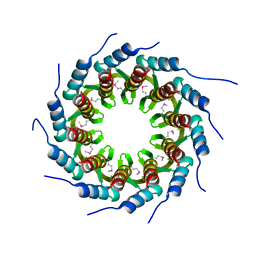 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V5I
 
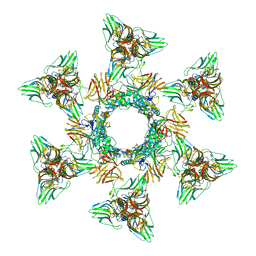 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3F0X
 
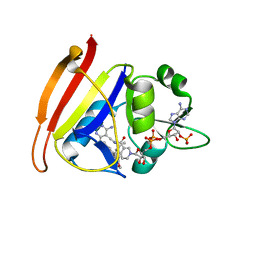 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-27 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Complexes of Wildtype and Mutant MRSA DHFR Reveal Interactions for Lead Design
To be Published
|
|
4UUS
 
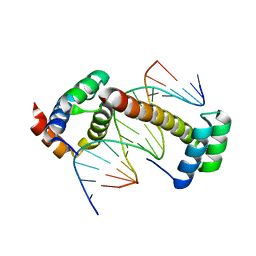 | | CRYSTAL STRUCTURE OF A UBX-EXD-DNA COMPLEX INCLUDING THE UBDA MOTIF | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*TP*TP*TP*AP*TP*GP*GP*CP*G)-3', 5'-D(*GP*TP*CP*GP*CP*CP*AP*TP*AP*AP*AP*TP*CP*AP*C)-3', HOMEOTIC PROTEIN EXTRADENTICLE, ... | | Authors: | Foos, N, Mate, M.J, Ortiz-Lombardia, M. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Flexible Extension of the Drosophila Ultrabithorax Homeodomain Defines a Novel Hox/Pbc Interaction Mode.
Structure, 23, 2015
|
|
4UUT
 
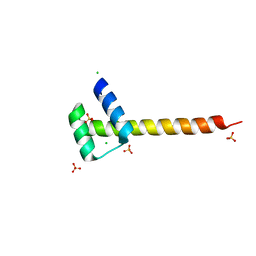 | |
1O6Y
 
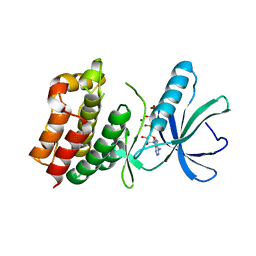 | | Catalytic domain of PknB kinase from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SERINE/THREONINE-PROTEIN KINASE PKNB | | Authors: | Ortiz-Lombardia, M, Pompeo, F, Boitel, B, Alzari, P.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Pknb Serine/Threonine Kinase from Mycobacterium Tuberculosis
J.Biol.Chem., 278, 2003
|
|
2CBY
 
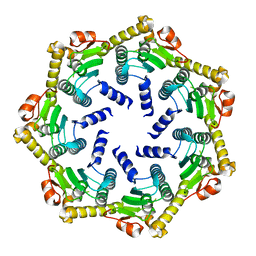 | | Crystal structure of the ATP-dependent Clp Protease proteolytic subunit 1 (ClpP1) from Mycobacterium tuberculosis | | Descriptor: | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 | | Authors: | Mate, M.J, Portnoi, D, Alzari, P.M, Ortiz-Lombardia, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into the Inter-Ring Plasticity of Caseinolytic Proteases from the X-Ray Structure of Mycobacterium Tuberculosis Clpp1.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2CE3
 
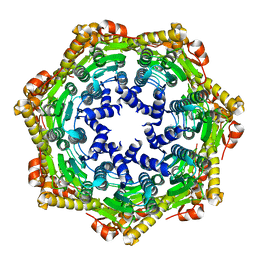 | | CRYSTAL STRUCTURE OF THE ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 (CLPP1) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 | | Authors: | Segelke, B, Kim, C.Y, Ortiz-Lombardia, M, Alzari, P.M, Lekin, T. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into the Inter-Ring Plasticity of Caseinolytic Proteases from the X-Ray Structure of Mycobacterium Tuberculosis Clpp1.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3U6X
 
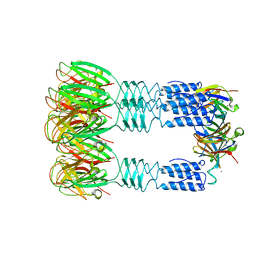 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2C5S
 
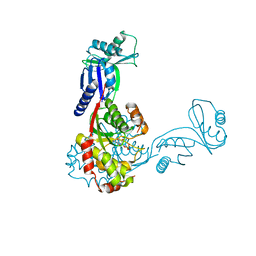 | | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROBABLE THIAMINE BIOSYNTHESIS PROTEIN THII | | Authors: | Waterman, D.G, Ortiz-Lombardia, M, Fogg, M.J, Koonin, E.V, Antson, A.A. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain.
J.Mol.Biol., 356, 2006
|
|
