8P2S
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/dTTP/GTP-bound state | | Descriptor: | Anaerobic ribonucleoside-triphosphate reductase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding.
Elife, 12, 2024
|
|
8P39
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-17 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding.
Elife, 12, 2024
|
|
8P2D
 
 | | Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding.
Elife, 12, 2024
|
|
1HK8
 
 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE, MANGANESE (II) ION, ... | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2003-03-06 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Metal-Binding Site in the Catalytic Subunit of Anaerobic Ribonucleotide Reductase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4YWT
 
 | |
2X40
 
 | |
3ZSJ
 
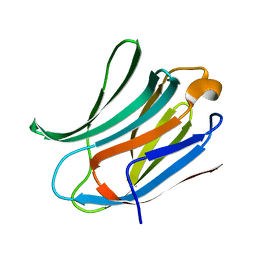 | | Crystal structure of Human Galectin-3 CRD in complex with Lactose at 0.86 angstrom resolution | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
7OLY
 
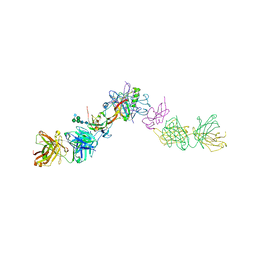 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
5K59
 
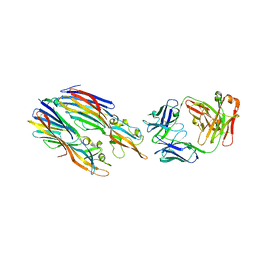 | | Crystal structure of LukGH from Staphylococcus aureus in complex with a neutralising antibody | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Welin, M, Logan, D.T, Badarau, A, Mirkina, I, Zauner, G, Dolezilkova, I, Nagy, E. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Mabs, 8, 2016
|
|
6FLH
 
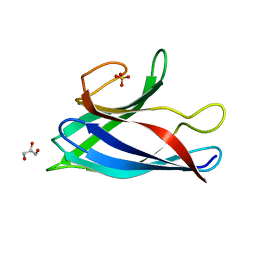 | | Monomeric Human Cu,Zn Superoxide dismutase, SOD1 7+7, apo form | | Descriptor: | GLYCEROL, SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Yang, F, Logan, D, Oliveberg, M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Cost of Long Catalytic Loops in Folding and Stability of the ALS-Associated Protein SOD1.
J.Am.Chem.Soc., 140, 2018
|
|
8P2C
 
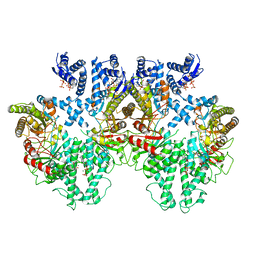 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric state produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding.
Elife, 12, 2024
|
|
6Y4C
 
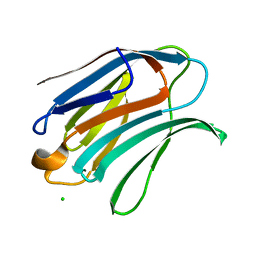 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using an XtalTool support | | Descriptor: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shilova, A, Hakansson, M, Welin, M, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
1BBT
 
 | | METHODS USED IN THE STRUCTURE DETERMINATION OF FOOT AND MOUTH DISEASE VIRUS | | Descriptor: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | Authors: | Acharya, K.R, Fry, E.E, Logan, D.T, Stuart, D.I. | | Deposit date: | 1992-05-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Methods used in the structure determination of foot-and-mouth disease virus.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
2BM1
 
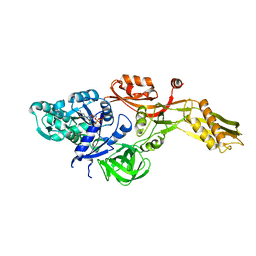 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant G16V | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
2BM0
 
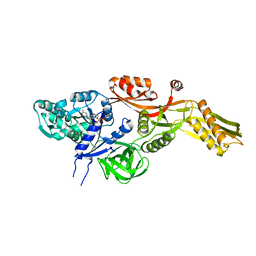 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant T84A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
6Y78
 
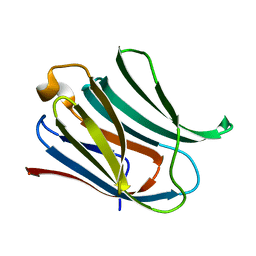 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using a silicon nitride membrane support | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hakansson, M, Welin, M, Shilova, A, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
2J7K
 
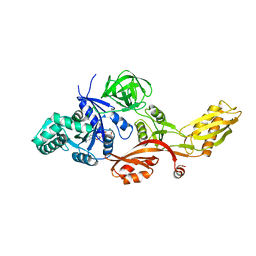 | | Crystal structure of the T84A mutant EF-G:GDPCP complex | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Hansson, S, Logan, D.T. | | Deposit date: | 2006-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New Insights Into the Role of the P-Loop Lysine: Implications from the Crystal Structure of a Mutant EF-G:Gdpcp Complex
To be Published
|
|
1H7B
 
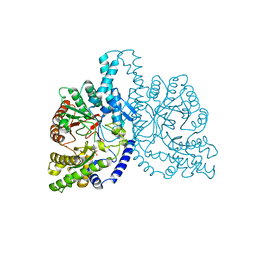 | | Structural basis for allosteric substrate specificity regulation in class III ribonucleotide reductases, native NRDD | | Descriptor: | ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, PHOSPHATE ION | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2001-07-04 | | Release date: | 2002-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
6EXY
 
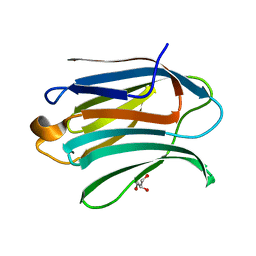 | | Neutron crystal structure of perdeuterated galectin-3C in complex with glycerol | | Descriptor: | GLYCEROL, Galectin-3 | | Authors: | Manzoni, F, Schrader, T.E, Ostermann, A, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-10 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
6F2Q
 
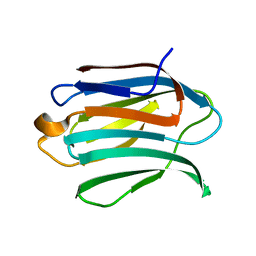 | | Neutron crystal structure of perdeuterated galectin-3C in the ligand-free form | | Descriptor: | Galectin-3 | | Authors: | Manzoni, F, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.03 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
7AMP
 
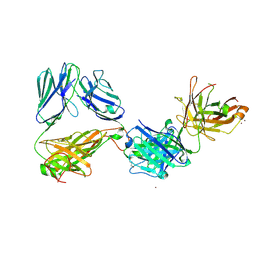 | | Crystal structure of the complex of HuJovi-1 Fab with the human A6 T-cell receptor TRBC1 | | Descriptor: | Alpha chain of A6 T-cell receptor, Beta chain 1 of A6 T-cell receptor TRBC1, CHLORIDE ION, ... | | Authors: | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
7AMQ
 
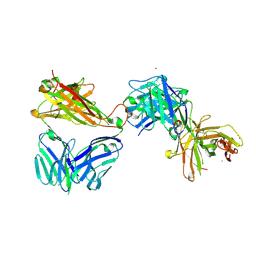 | | Crystal structure of the complex of HuJovi-1 Fab with the human TRBC2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Human A6 T-cell receptor alpha chain, ... | | Authors: | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
7AMS
 
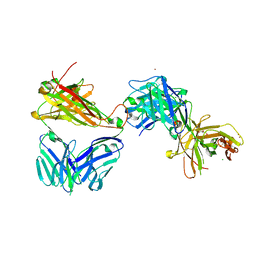 | | Crystal structure of the complex of the KFN mutant of HuJovi-1 Fab with human TRBC2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Human A6 T-cell receptor alpha chain, ... | | Authors: | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Svensson, A, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
7AMR
 
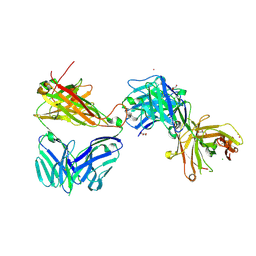 | | Crystal structure of the complex of the KFN mutant of Jovi-1 Fab with human TRBC1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Human Jovi-1 Fab fragment, ... | | Authors: | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
7BFG
 
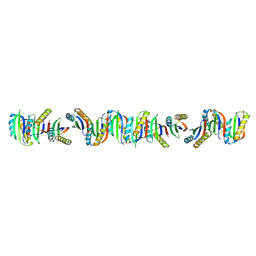 | |
