2AJH
 
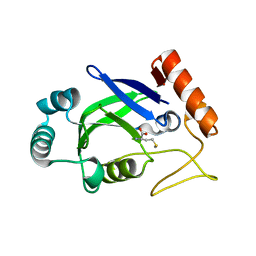 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with methionine | | Descriptor: | Leucyl-tRNA synthetase, METHIONINE | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2AJI
 
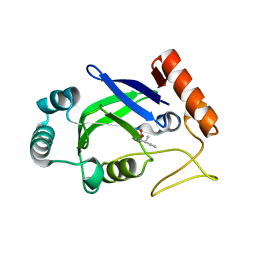 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with isoleucine | | Descriptor: | ISOLEUCINE, Leucyl-tRNA synthetase | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2AZP
 
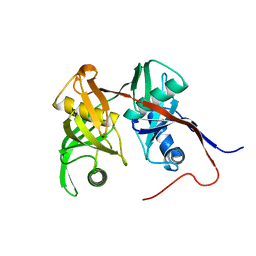 | | Crystal Structure of PA1268 Solved by Sulfur SAD | | Descriptor: | hypothetical protein PA1268 | | Authors: | Liu, Y, Gorodichtchenskaia, E, Skarina, T, Yang, C, Joachimiak, A, Edwards, A, Pai, E.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of PA1268 Solved by Sulfur SAD
To be Published
|
|
5F3W
 
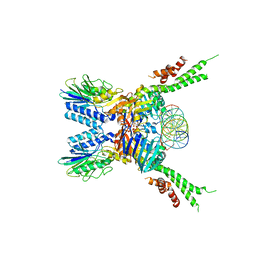 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | 27-MER DNA, DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Embo J., 35, 2016
|
|
2EAW
 
 | |
3MO7
 
 | |
2ESH
 
 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
3MI2
 
 | | Crystal structure of human orotidine-5'-monophosphate decarboxylase complexed with pyrazofurin monophosphate | | Descriptor: | (1S)-1,4-anhydro-1-(5-carbamoyl-4-hydroxy-1H-pyrazol-3-yl)-5-O-phosphono-D-ribitol, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, To, T, Kotra, L.P, Pai, E.F. | | Deposit date: | 2010-04-09 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural determinants for the inhibitory ligands of orotidine-5'-monophosphate decarboxylase.
Bioorg.Med.Chem., 18, 2010
|
|
3MW7
 
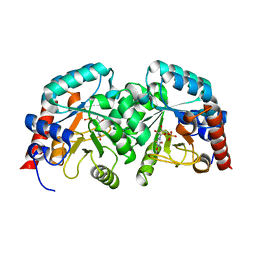 | |
2FRH
 
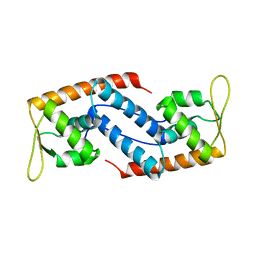 | | Crystal Structure of Sara, A Transcription Regulator From Staphylococcus Aureus | | Descriptor: | CALCIUM ION, Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Ingavale, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FNP
 
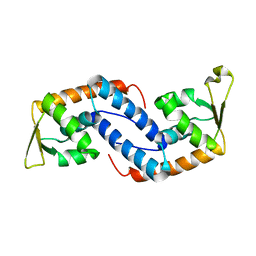 | | Crystal structure of SarA | | Descriptor: | Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Pan, C.H, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6ASG
 
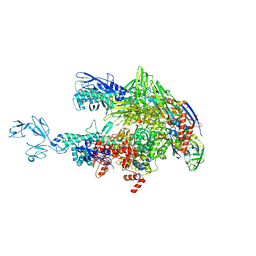 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
4PLI
 
 | |
4PLL
 
 | |
4RGY
 
 | |
3BGJ
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP | | Descriptor: | GLYCEROL, URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Devalla, S, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP
To be Published
|
|
3BGG
 
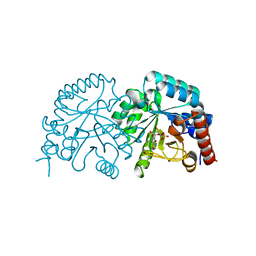 | | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Wang, X.Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP
To be Published
|
|
4O6X
 
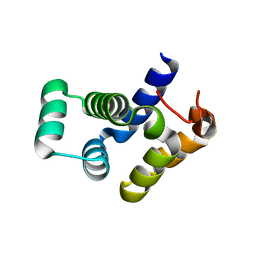 | |
4Q46
 
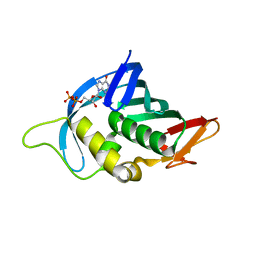 | |
3N2M
 
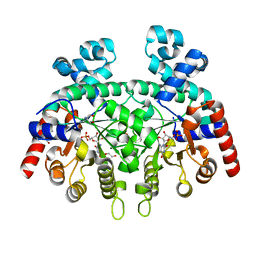 | | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2010-05-18 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP
To be Published
|
|
3N34
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP, produced from 5-fluoro-6-azido-UMP | | Descriptor: | 1,2-ETHANEDIOL, 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2010-05-19 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP, produced from 5-fluoro-6-azido-UMP
To be Published
|
|
3MWA
 
 | |
3N3M
 
 | |
3PST
 
 | |
3PSP
 
 | |
