3SR7
 
 | |
3SQZ
 
 | |
8AHL
 
 | |
8AFH
 
 | |
8AJB
 
 | |
8AIA
 
 | |
8AIX
 
 | |
8AFE
 
 | |
8AFM
 
 | |
8AFL
 
 | |
3G3M
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-iodo-UMP | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Poduch, E, Kotra, L.P, Pai, E.F. | | Deposit date: | 2009-02-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
8GO5
 
 | |
8GO7
 
 | | Fungal immunomodulatory protein FIP-nha N5+39A | | Descriptor: | Fungal immunomodulatory protein FIP-nha | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
7RDQ
 
 | |
3TEJ
 
 | |
2KJP
 
 | | Solution structure of protein YlbL (BSU15050) from Bacillus subtilis, Northeast Structural Genomics Consortium target sr713a | | Descriptor: | Uncharacterized protein ylbL | | Authors: | Liu, Y, Belote, R, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T, Xiao, R, Swapna, G, Everett, J, Montelione, G.T, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein YlbL (BSU15050) from Bacillus subtilis, Northeast Structural Genomics Consortium target sr713a
To be Published
|
|
8GO6
 
 | | Fungal immunomodulatory protein FIP-nha N39A | | Descriptor: | fungal immunomodulatory protein FIP-nha N39A | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
3THQ
 
 | |
6X9H
 
 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
3SW6
 
 | |
1JQ1
 
 | |
6V2S
 
 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2R
 
 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
1JQ2
 
 | |
1JS0
 
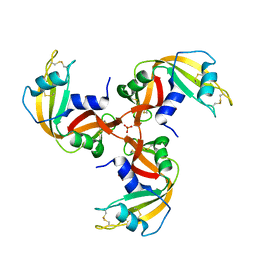 | | Crystal Structure of 3D Domain-swapped RNase A Minor Trimer | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Gotte, G, Libonati, M, Eisenberg, D. | | Deposit date: | 2001-08-15 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the two 3D domain-swapped RNase A trimers.
Protein Sci., 11, 2002
|
|
