4BEV
 
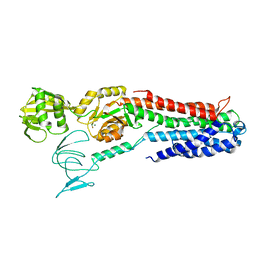 | | ATPase crystal structure with bound phosphate analogue | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, TRIFLUOROMAGNESATE | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Gourdon, P, Pedersen, B.P, Morth, P, Wang, J, Nissen, P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | ATPase Crystal Structure with Bound Phosphate Analogue
To be Published
|
|
3J0C
 
 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
7JXG
 
 | | Structural model for Fe-containing human acireductone dioxygenase | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, FE (II) ION | | Authors: | Pochapsky, T.C, Liu, X, Deshpande, A, Ringe, D, Garber, A, Ryan, J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).
Biochemistry, 59, 2020
|
|
8Q4L
 
 | | GBP1 bound by 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, Guanylate-binding protein 1 | | Authors: | Pfleiderer, M.M, Liu, X, Fisch, D, Anastasakou, E, Frickel, E.M, Galej, W.P. | | Deposit date: | 2023-08-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (5.12 Å) | | Cite: | PIM1 controls GBP1 activity to limit self-damage and to guard against pathogen infection.
Science, 382, 2023
|
|
1Q2R
 
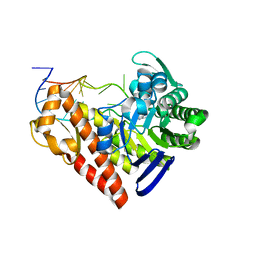 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, RNA (5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(N)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3'), ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
1PUY
 
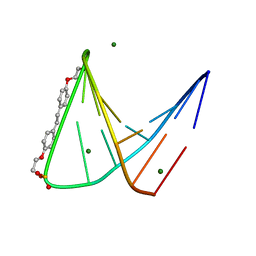 | | 1.5 A resolution structure of a synthetic DNA hairpin with a stilbenediether linker | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*GP*(S02)P*CP*AP*AP*AP*AP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Murshudov, G, Sanishvili, R, Liu, X, Lewis, F.D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
J.Am.Chem.Soc., 125, 2003
|
|
5T4D
 
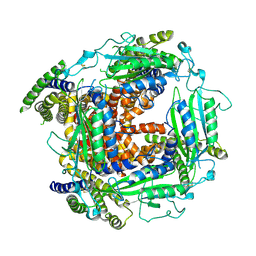 | | Cryo-EM structure of Polycystic Kidney Disease protein 2 (PKD2), residues 198-703 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hPKD:198-703, Polycystin-2 | | Authors: | Shen, P.S, Yang, X, DeCaen, P.G, Liu, X, Bulkley, D, Clapham, D.E, Cao, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs.
Cell, 167, 2016
|
|
1Q2S
 
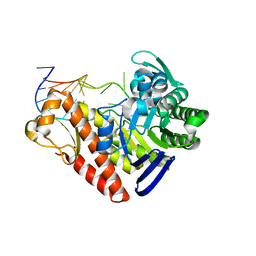 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(PQ1)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3', 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
2F7X
 
 | | Protein Kinase A bound to (S)-2-(1H-Indol-3-yl)-1-[5-((E)-2-pyridin-4-yl-vinyl)-pyridin-3-yloxymethyl]-ethylamine | | Descriptor: | (1S)-2-(1H-INDOL-3-YL)-1-[({5-[(E)-2-PYRIDIN-4-YLVINYL]PYRIDIN-3-YL}OXY)METHYL]ETHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Li, T, Zhu, G.D, Gong, J, Claibone, A, Dalton, C, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Bauch, J.L, Marsh, K.C, Bouska, J.J, Arries, S, De Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of trans-3,4'-bispyridinylethylenes as potent and novel inhibitors of protein kinase B (PKB/Akt) for the treatment of cancer: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
9IJD
 
 | | Carazolol-activated human beta3 adrenergic receptor | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, Beta-3 adrenergic receptor, Camelid antibody VHH fragment, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular Mechanism of the beta 3AR Agonist Activity of a beta-Blocker.
Chempluschem, 2024
|
|
9IJE
 
 | | Epinephrine-activated human beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Camelid antibody VHH fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular Mechanism of the beta 3AR Agonist Activity of a beta-Blocker.
Chempluschem, 2024
|
|
4EQA
 
 | | Crystal structure of PA1844 in complex with PA1845 from Pseudomonas aeruginosa PAO1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
7MNJ
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
3L82
 
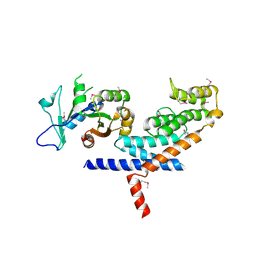 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
8ED0
 
 | |
7MVW
 
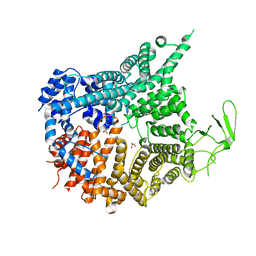 | | Crystal structure of Chaetomium thermophilum Nup188 NTD (residues 1-1134) | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVT
 
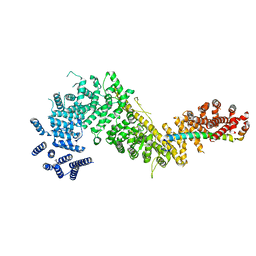 | | Crystal structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 185-1756; Nic96 residues 187-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
3J7L
 
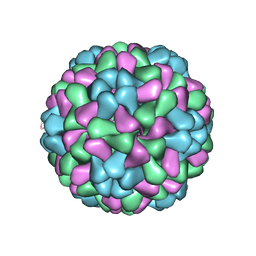 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
2VAS
 
 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
2VB6
 
 | | Myosin VI (MD-insert2-CaM, Delta Insert1) Post-rigor state (crystal form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
