5XH3
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with HEMT from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, O 4-(2-hydroxyethyl) O 1-methyl benzene-1,4-dicarboxylate, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
4EN0
 
 | | Crystal structure of light | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhan, C, Liu, W, Patskovsky, Y, Ramagopal, U.A, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
8GPD
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxidanyl-2-oxidanylidene-1-(2-phenoxyethanoylamino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPC
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, SODIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPE
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin G | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
5YNW
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP and INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
7Y3Z
 
 | |
7Y9J
 
 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium in complex with 5-nitro-1,2-benzisoxazole | | Descriptor: | 5-nitro-1,2-benzoxazole, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
7Y9K
 
 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
4FVD
 
 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
5YGJ
 
 | | Crystal structure of a synthase from Streptomyces sp. CL190 | | Descriptor: | Cyclolavandulyl diphosphate synthase | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
4FVB
 
 | | Crystal structure of EV71 2A proteinase C110A mutant | | Descriptor: | 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
7V40
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, p450tol monooxygenase | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V41
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with toluene. | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, TOLUENE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V44
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with meta-chlorotoluene. | | Descriptor: | 1-chloranyl-2-methyl-benzene, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V43
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with para-chlorotoluene. | | Descriptor: | 1-chloranyl-4-methyl-benzene, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V42
 
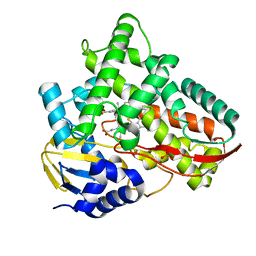 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with benzyl-alcohol. | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, p450tol monooxygenase, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V45
 
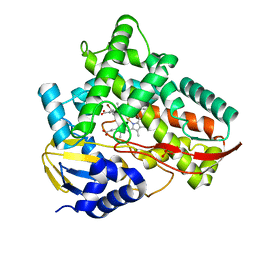 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with para-bromotoluene. | | Descriptor: | 1-bromanyl-4-methyl-benzene, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V46
 
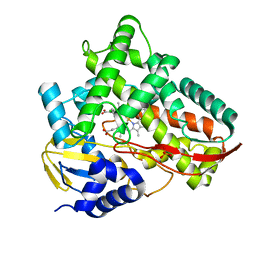 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with ortho-chlorotoluene. | | Descriptor: | 1-chloranyl-3-methyl-benzene, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
6JJS
 
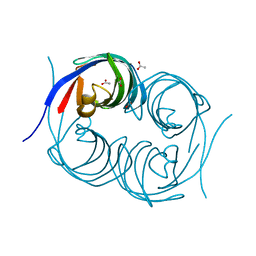 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
5YNT
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
6JJT
 
 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
5YNU
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YNV
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, aromatic prenyltransferase | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
