7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
4WXO
 
 | |
8ZFK
 
 | | Caenorhabditis elegans ACR-23 in betaine and monepantel bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betaine receptor acr-23,Soluble cytochrome b562, ... | | Authors: | Chen, Q.F, Liu, F.L, Li, T.Y, Gong, H.H, Guo, F, Liu, S. | | Deposit date: | 2024-05-08 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural basis of acetylcholine receptor like-23 (ACR-23) activation by anthelmintics monepantel and betaine
To Be Published
|
|
1FAW
 
 | | GRAYLAG GOOSE HEMOGLOBIN (OXY FORM) | | Descriptor: | HEMOGLOBIN (ALPHA SUBUNIT), HEMOGLOBIN (BETA SUBUNIT), OXYGEN MOLECULE, ... | | Authors: | Liang, Y.-H, Liu, X.-Z, Liu, S.-H, Lu, G.-Y. | | Deposit date: | 2000-07-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The structure of greylag goose oxy haemoglobin: the roles of four mutations compared with bar-headed goose haemoglobin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5WIX
 
 | | Crystal structure of P47 of Clostridium botulinum E1 | | Descriptor: | p-47 protein | | Authors: | Lam, K, Liu, S, Qi, R, Jin, R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein.
Toxicon, 147, 2018
|
|
4WXW
 
 | |
6WVG
 
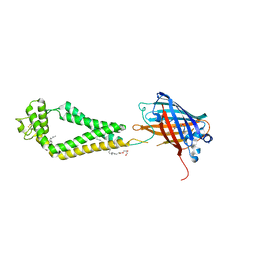 | | human CD53 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein, ... | | Authors: | Yang, Y, Liu, S, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Open conformation of tetraspanins shapes interaction partner networks on cell membranes.
Embo J., 39, 2020
|
|
1ST0
 
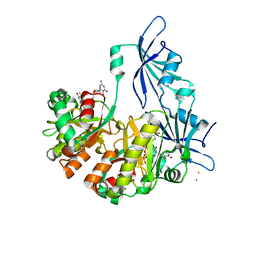 | | Structure of DcpS bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
1ST4
 
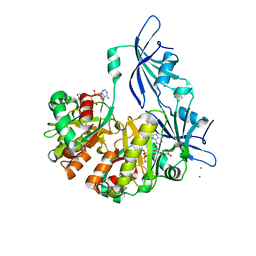 | | Structure of DcpS bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
8YF1
 
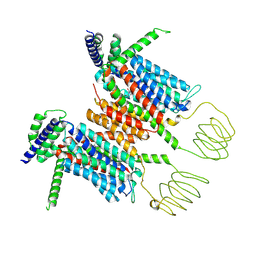 | |
8YF0
 
 | |
4IT7
 
 | | Crystal structure of Al-CPI | | Descriptor: | CPI | | Authors: | Mei, G.Q, Liu, S.L, Sun, M.Z, Liu, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-01-29 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Immunomodulatory Function of Cysteine Protease Inhibitor from Human Roundworm Ascaris lumbricoides.
Plos One, 9, 2014
|
|
4ZMU
 
 | | Dcsbis, a diguanylate cyclase from Pseudomonas aeruginosa | | Descriptor: | diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | crystal structure of Dcsbis from Pseudomonas aeruginosa
To Be Published
|
|
4ZMM
 
 | | GGDEF domain of Dcsbis complexed with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of Dcsbis GGDEF domain complexed with c-di-GMP
To Be Published
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | Descriptor: | Endolysin, PHENYLETHANE | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWT
 
 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | Descriptor: | BENZENE, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWU
 
 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
7QPT
 
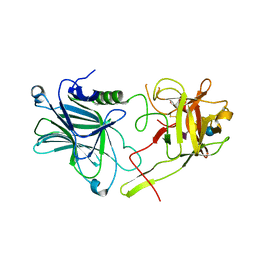 | | Botulinum neurotoxin A4 cell binding domain in complex with GD1a oligosaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type A | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mojanaga, O.O. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
3QIC
 
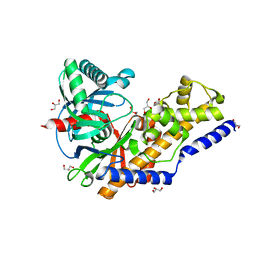 | | The structure of human glucokinase E339K mutation | | Descriptor: | GLYCEROL, Glucokinase, alpha-D-glucopyranose | | Authors: | Liu, Q, Liu, S, Liu, J. | | Deposit date: | 2011-01-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of E339K mutated human glucokinase reveals changes in the ATP binding site.
Febs Lett., 585, 2011
|
|
7QPU
 
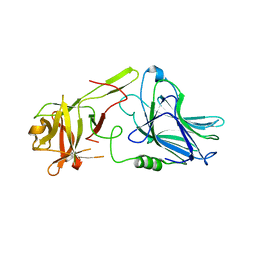 | | Botulinum neurotoxin A5 cell binding domain in complex with GM1b oligosaccharide | | Descriptor: | Botulinum neurotoxin sub-type A5, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
4R0J
 
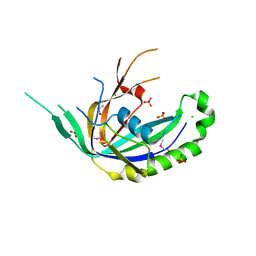 | | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans
To be Published
|
|
6F0P
 
 | | Botulinum neurotoxin A4 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, J.R, Rees, J, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
6F0O
 
 | | Botulinum neurotoxin A3 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, Bontoxilysin A, PENTAETHYLENE GLYCOL, ... | | Authors: | Davies, J.R, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
