1V6P
 
 | | Crystal structure of Cobrotoxin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Cobrotoxin, ... | | Authors: | Lou, X, Tu, X, Wang, J, Teng, M, Niu, L, Liu, Q, Huang, Q, Hao, Q. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
1TW9
 
 | | Glutathione Transferase-2, apo form, from the nematode Heligmosomoides polygyrus | | Descriptor: | glutathione S-transferase 2 | | Authors: | Schuller, D.J, Liu, Q, Kriksunov, I.A, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of a new class of glutathione transferase from the model human hookworm nematode Heligmosomoides polygyrus.
Proteins, 61, 2005
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | Deposit date: | 2022-01-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
2ITG
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
1XPQ
 
 | | Crystal structure of fms1, a polyamine oxidase from yeast | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, SPERMINE | | Authors: | Huang, Q, Liu, Q, Hao, Q. | | Deposit date: | 2004-10-09 | | Release date: | 2005-04-26 | | Last modified: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
J.Mol.Biol., 348, 2005
|
|
1YY5
 
 | |
6PBR
 
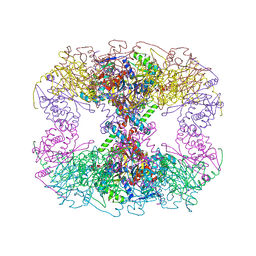 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | Authors: | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NQ8
 
 | |
6NQ9
 
 | |
6NQ7
 
 | |
4U9A
 
 | |
8FT4
 
 | | Multicrystal structure of Na+, leucine-bound LeuT determined at 5 keV | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Karasawa, A, Liu, H, Quick, M, Hendrickson, A.H, Liu, Q. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystallographic characterization of sodium ions in a bacterial leucine/sodium symporter
To be Published
|
|
8FT5
 
 | | Crystal structure of LeuT soaked with Crown-5 | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Karasawa, A, Liu, H, Quick, M, Hendrickson, A.H, Liu, Q. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystallographic characterization of sodium ions in a bacterial leucine/sodium symporter
To be Published
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
5K0X
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
1ZL9
 
 | | Crystal Structure of a major nematode C.elegans specific GST (CE01613) | | Descriptor: | GLUTATHIONE, glutathione S-transferase 5 | | Authors: | Kriksunov, I.A, Liu, Q, Schuller, D.J, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a major nematode C.elegans specific GST (CE01613)
To be Published
|
|
4WD7
 
 | |
2QXL
 
 | | Crystal Structure Analysis of Sse1, a yeast Hsp110 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein homolog SSE1, MAGNESIUM ION, ... | | Authors: | Hendrickson, W.A, Liu, Q. | | Deposit date: | 2007-08-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insights into hsp70 chaperone activity from a crystal structure of the yeast hsp110 Sse1.
Cell(Cambridge,Mass.), 131, 2007
|
|
4JN4
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3I9O
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Graeff, R, Liu, Q, Kriksunov, I.A, Kotaka, M, Oppenheimer, N, Hao, Q, Lee, H.C. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38
J.Biol.Chem., 284, 2009
|
|
