3CFU
 
 | | Crystal structure of the yjhA protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR562 | | Descriptor: | Uncharacterized lipoprotein yjhA | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Forouhar, F, Zhao, L, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Swapna, G, Huang, J.Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the yjhA protein from Bacillus subtilis.
To be Published
|
|
3CET
 
 | | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | Conserved archaeal protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, J.F, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63.
To be Published
|
|
3CER
 
 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | Descriptor: | Possible exopolyphosphatase-like protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
5V9J
 
 | | Crystal structure of catalytic domain of GLP with MS0105 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of GLP with MS0105
to be published
|
|
4IU6
 
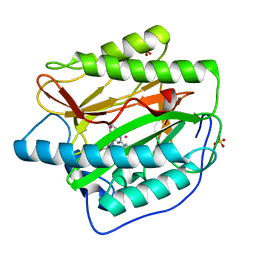 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | Descriptor: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | Deposit date: | 2013-01-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
3D0W
 
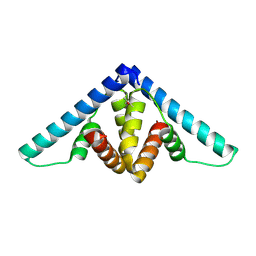 | | Crystal structure of YflH protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR326 | | Descriptor: | YflH protein | | Authors: | Seetharaman, J, Kuzin, A.P, Neely, H, Forouhar, F, Min, S, Zhao, L, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YflH protein from Bacillus subtilis.
To be Published
|
|
3D3E
 
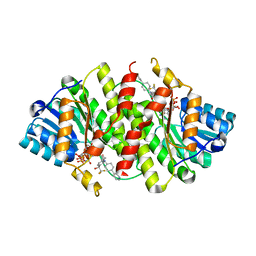 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Benzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-cyclopropyl-N-(trans-4-pyridin-3-ylcyclohexyl)-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Liu, J, Walker, N.P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
5VSC
 
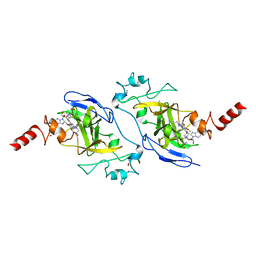 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 13 | | Descriptor: | 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
3D5Q
 
 | | Crystal Structure of 11b-HSD1 in Complex with Triazole Inhibitor | | Descriptor: | 3-[1-(4-fluorophenyl)cyclopropyl]-4-(1-methylethyl)-5-[4-(trifluoromethoxy)phenyl]-4H-1,2,4-triazole, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P.C. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Distinctive molecular inhibition mechanisms for selective inhibitors of human 11beta-hydroxysteroid dehydrogenase type 1.
Bioorg.Med.Chem., 16, 2008
|
|
7SZE
 
 | |
7SZH
 
 | |
7SZG
 
 | | Structure of the Rieske Non-heme Iron Oxygenase GxtA Pressurized with Xenon | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Bridwell-Rabb, J, Liu, J. | | Deposit date: | 2021-11-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design principles for site-selective hydroxylation by a Rieske oxygenase.
Nat Commun, 13, 2022
|
|
7SZF
 
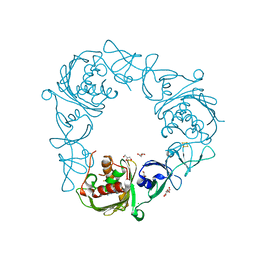 | |
3CKZ
 
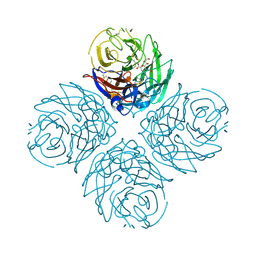 | | N1 Neuraminidase H274Y + Zanamivir | | Descriptor: | CALCIUM ION, Neuraminidase, ZANAMIVIR | | Authors: | Colllins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3D4N
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Sulfonamide Inhibitor | | Descriptor: | 1-{[(3R)-3-methyl-4-({4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]phenyl}sulfonyl)piperazin-1-yl]methyl}cyclopropanecarboxamide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P. | | Deposit date: | 2008-05-14 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
5VZL
 
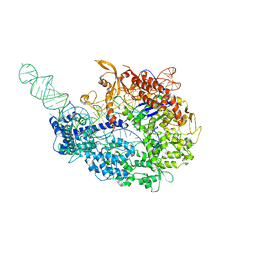 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
5TTG
 
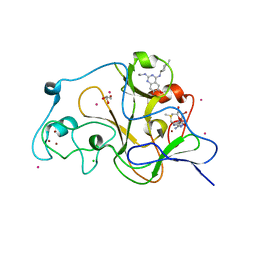 | | Crystal structure of catalytic domain of GLP with MS012 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
2ICT
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSE
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 17: N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
3CL2
 
 | | N1 Neuraminidase N294S + Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
2INW
 
 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3FHW
 
 | | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Primosomal replication protein n, SODIUM ION | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162.
To be Published
|
|
