3P40
 
 | |
2O26
 
 | | Structure of a class III RTK signaling assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor, ... | | Authors: | Liu, H, Chen, X, Focia, P.J, He, X. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases.
Embo J., 26, 2007
|
|
4JQE
 
 | | Crystal structure of scCK2 alpha in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3P3Y
 
 | |
4JR7
 
 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | Descriptor: | Casein kinase II subunit alpha, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG5
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with oxaloacetate | | Descriptor: | CACODYLATE ION, GLYCEROL, OXALOACETATE ION, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5ZVT
 
 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | Descriptor: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | Authors: | Liu, H, Fang, Q, Cheng, L. | | Deposit date: | 2018-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5FBJ
 
 | | Complex structure of JMJD5 and substrate | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, ... | | Authors: | Liu, H.L, Wang, Y, Wang, C, Zhang, G.Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | to be published
To Be Published
|
|
4HGD
 
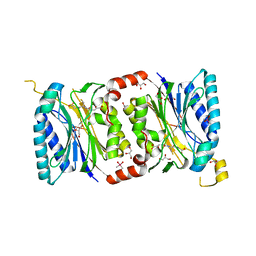 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG3
 
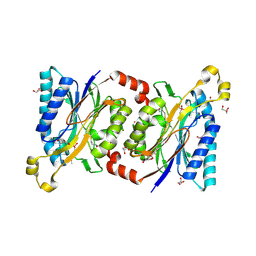 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C1R
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist avacopan | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MALONATE ION, OLEIC ACID, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7D32
 
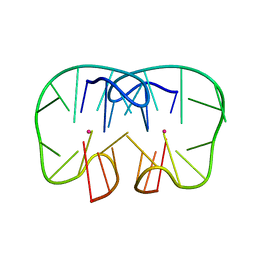 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D33
 
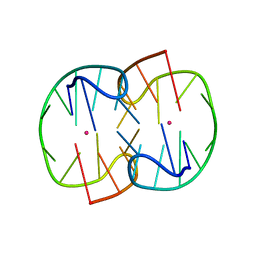 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
4LFI
 
 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | Descriptor: | Casein kinase II subunit alpha, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Liu, H. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MWH
 
 | | Crystal structure of scCK2 alpha in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Liu, H. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8J19
 
 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1A
 
 | | Cryo-EM structure of the GPR84 receptor-Gi complex with no ligand modeled | | Descriptor: | Antibody fragment ScFv16, G-protein coupled receptor 84, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
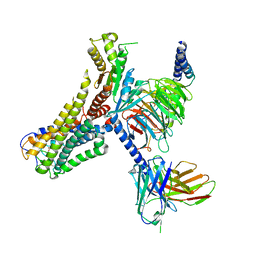 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
5ZVS
 
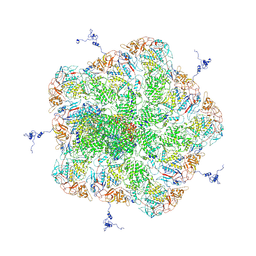 | |
7D31
 
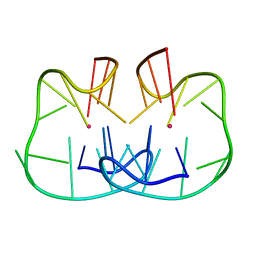 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
5XMA
 
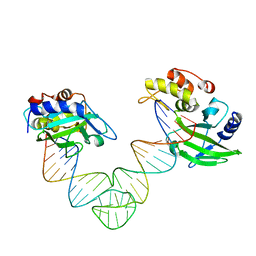 | |
2O27
 
 | |
8H89
 
 | | Capsid of Ralstonia phage GP4 | | Descriptor: | Major capsid protein, Virion associated protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-10-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.
Viruses, 14, 2022
|
|
