3JA5
 
 | |
8I4M
 
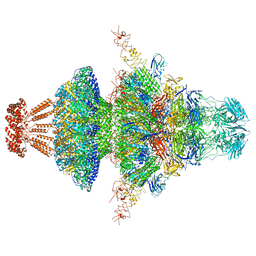 | | Portal-tail complex structure of the Cyanophage P-SCSP1u | | Descriptor: | Adaptor protein(gp22) of the cyanophage P-SCSP1u, Fiber protein(gp 28) of the cyanophage P-SCSP1u, Nozzle protein(gp 23) of the cyanophage P-SCSP1u, ... | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
5BT1
 
 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | Descriptor: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | Authors: | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
5XK0
 
 | | Structure of 8-mer DNA2 | | Descriptor: | DNA (5'-D(*GP*CP*CP*CP*GP*AP*GP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XK1
 
 | | Structure of 8-mer DNA3 | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*CP*CP*CP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4JQE
 
 | | Crystal structure of scCK2 alpha in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JR7
 
 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | Descriptor: | Casein kinase II subunit alpha, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HGD
 
 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG3
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG5
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with oxaloacetate | | Descriptor: | CACODYLATE ION, GLYCEROL, OXALOACETATE ION, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5Y4M
 
 | | Discoidin domain of human CASPR2 | | Descriptor: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | Authors: | Liu, H, Xu, F, Zhang, J, Liang, W. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
4MWH
 
 | | Crystal structure of scCK2 alpha in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Liu, H. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KTR
 
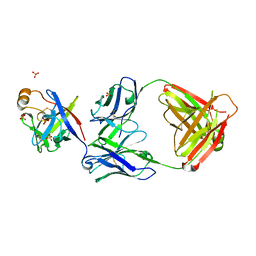 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
7JMX
 
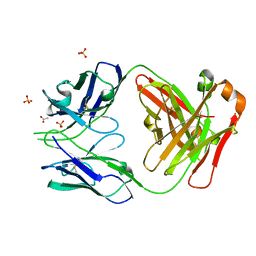 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
4QU2
 
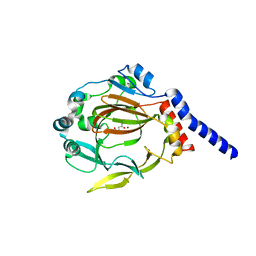 | | The structure of JMJD7 with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, JmjC domain-containing protein 7, NICKEL (II) ION | | Authors: | Liu, H.L, Wang, C, Zhang, G.Y. | | Deposit date: | 2014-07-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of JMJD7 with alpha-KG
To be Published
|
|
7D33
 
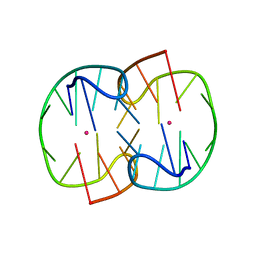 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D32
 
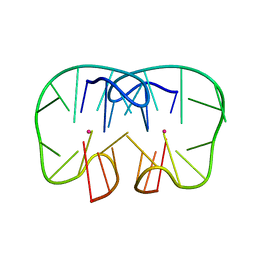 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
5ZVS
 
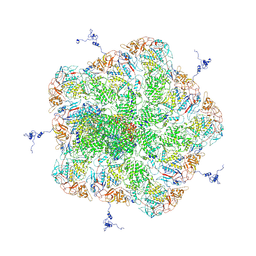 | |
7JMW
 
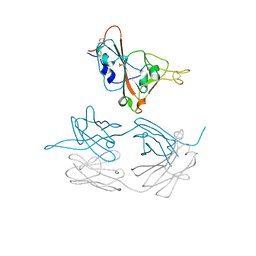 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
3NTI
 
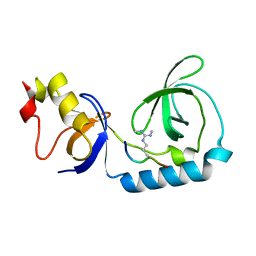 | | Crystal structure of Tudor and Aubergine [R15(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
3NTK
 
 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
3NTH
 
 | | Crystal structure of Tudor and Aubergine [R13(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
7D31
 
 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
3P40
 
 | |
