7YVH
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH132 Fab | | Descriptor: | Spike glycoprotein, TH132 Fab heavy chain, TH132 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVM
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH281 Fab heavy chain, TH281 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVI
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH236 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVL
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH272 Fab heavy chain, TH272 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVJ
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH236 Fab | | Descriptor: | Spike glycoprotein, TH236 Fab heavy chain, TH236 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
4X8J
 
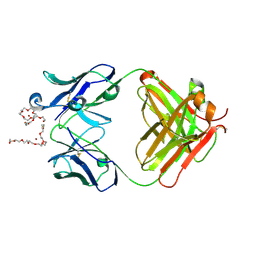 | | Crystal Structure of murine 12F4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | 12F4 FAB Heavy chain, 12F4 FAB Light chain, NONAETHYLENE GLYCOL, ... | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
1XAX
 
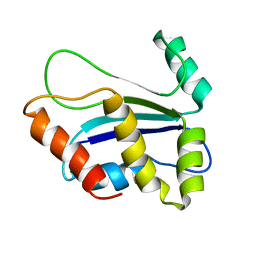 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
7YI8
 
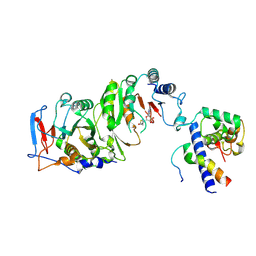 | | Cryo-EM structure of SAH-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
7YI9
 
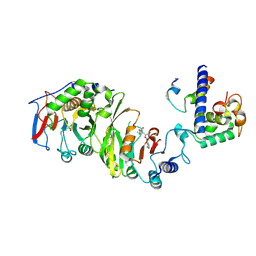 | | Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
3OZP
 
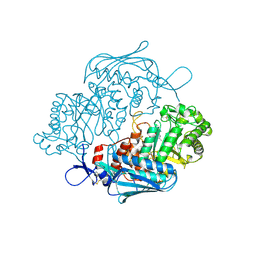 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases
Biochem.J., 438, 2011
|
|
4X80
 
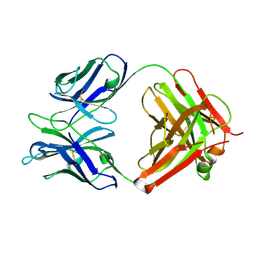 | | Crystal Structure of murine 7B4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | IgG1 7B4 FAB Heavy chain, IgG1 7B4 FAB Light Chain | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
3NSM
 
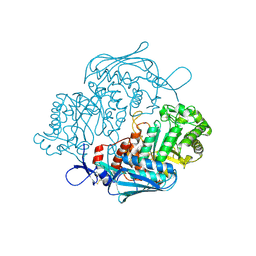 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
2AOF
 
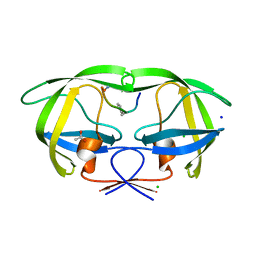 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P1-P6 | | Descriptor: | ACETIC ACID, CHLORIDE ION, PEPTIDE INHIBITOR, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | Descriptor: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOE
 
 | | crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog CA-P2 | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOH
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | Descriptor: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
3OZO
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
2AOC
 
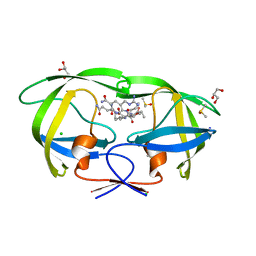 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOD
 
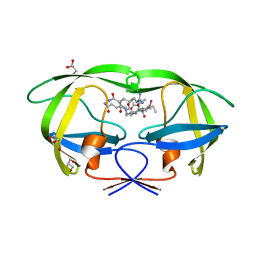 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
4OFC
 
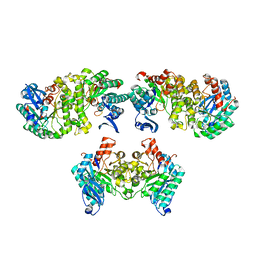 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
8JR0
 
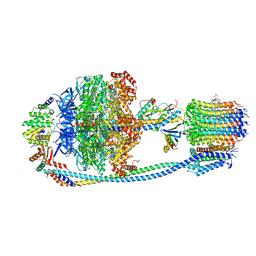 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
2ELA
 
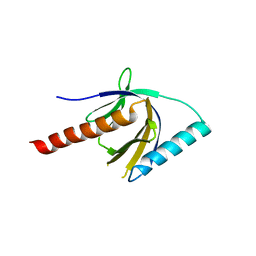 | | Crystal Structure of the PTB domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
4OE2
 
 | | 2.00 Angstroms X-ray crystal structure of E268A 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Esaki, S, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
