8B1R
 
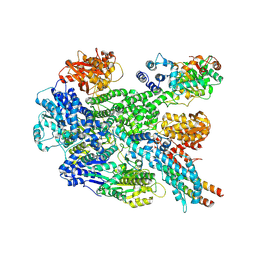 | | RecBCD in complex with the phage protein gp5.9 | | Descriptor: | MAGNESIUM ION, Probable RecBCD inhibitor gp5.9, RecBCD enzyme subunit RecB, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
8B1T
 
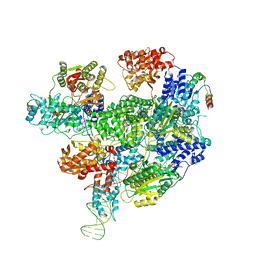 | | RecBCD-DNA in complex with the phage protein Abc2 | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
6Y3H
 
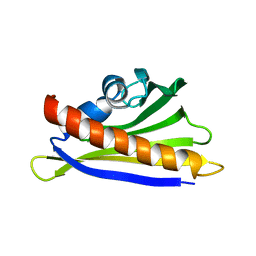 | | NMR solution structure of the hazelnut allergen Cor a 1.0401 | | Descriptor: | Major allergen Cor a 1.0401 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3L
 
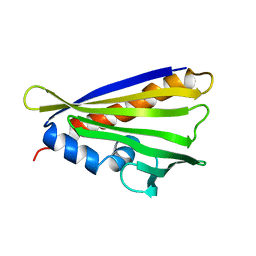 | | NMR solution structure of the hazelnut allergen Cor a 1.0404 | | Descriptor: | Major allergen variant Cor a 1.0404 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3I
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0402 | | Descriptor: | Major allergen variant Cor a 1.0402 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3K
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | Descriptor: | Major allergen variant Cor a 1.0403 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Z98
 
 | | NMR solution structure of the peach allergen Pru p 1.0101 | | Descriptor: | Major allergen Pru p 1 | | Authors: | Eidelpes, R, Fuehrer, S, Hofer, F, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Zeatin Binding of the Peach Allergen Pru p 1 .
J.Agric.Food Chem., 69, 2021
|
|
5MMU
 
 | | NMR solution structure of the major apple allergen Mal d 1 | | Descriptor: | Major allergen Mal d 1 | | Authors: | Ahammer, L, Grutsch, S, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major Apple Allergen Mal d 1.
J. Agric. Food Chem., 65, 2017
|
|
5DMA
 
 | |
5N5C
 
 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
5NOC
 
 | |
5LYM
 
 | |
5ANY
 
 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
4IFJ
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
4IFN
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4IFL
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
5AQB
 
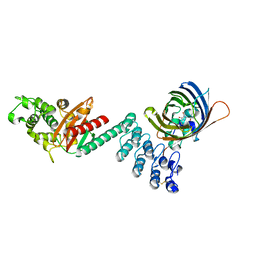 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 3G61_DB15V4, GREEN FLUORESCENT PROTEIN | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQ9
 
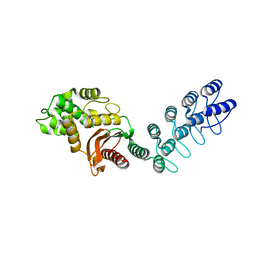 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, OFF7_DB08V4 | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
3V3M
 
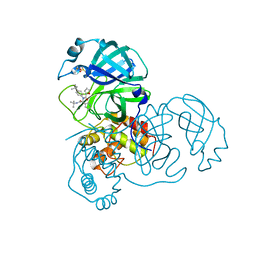 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
2YII
 
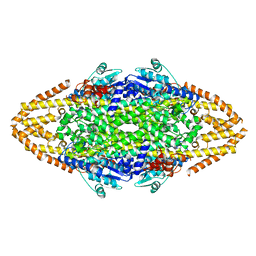 | | Manipulating the regioselectivity of phenylalanine aminomutase: new insights into the reaction mechanism of MIO-dependent enzymes from structure-guided directed evolution | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Szymanski, W, Wybenga, G.G, Heberling, M.M, Bartsch, S, Wildeman, S, Poelarends, G.J, Feringa, B.L, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism-Inspired Engineering of Phenylalanine Aminomutase for Enhanced Beta-Regioselective Asymmetric Amination of Cinnamates.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7VRT
 
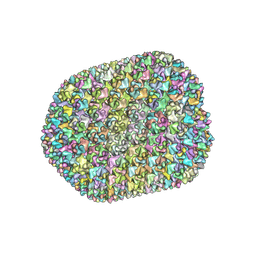 | | The unexpanded head structure of phage T4 | | Descriptor: | Capsid vertex protein, Major capsid protein | | Authors: | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VS5
 
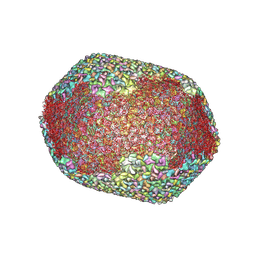 | | The expanded head structure of phage T4 | | Descriptor: | Capsid vertex protein, Major capsid protein, Small outer capsid protein | | Authors: | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2021-10-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2YNP
 
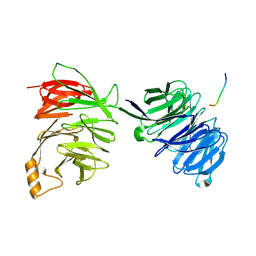 | | yeast betaprime COP 1-604 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.962 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2YNN
 
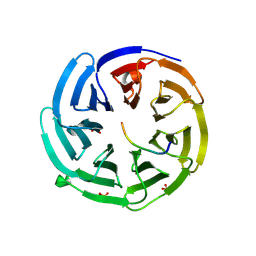 | | yeast betaprime COP 1-304 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF, SULFATE ION | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
4N4Z
 
 | | Trypanosoma brucei procathepsin B structure solved by Serial Microcrystallography using synchrotron radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gati, C, Bourenkov, G, Klinge, M, Rehders, D, Stellato, F, Oberthuer, D, White, T.A, Yevanov, O, Sommer, B.P, Mogk, S, Duszenko, M, Betzel, C, Schneider, T.R, Chapman, H.N, Redecke, L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Serial crystallography on in vivo grown microcrystals using synchrotron radiation.
IUCrJ, 1, 2014
|
|
