2RDJ
 
 | |
2IO4
 
 | | Crystal structure of PCNA12 dimer from Sulfolobus solfataricus. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DNA polymerase sliding clamp B, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-10-09 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2IA6
 
 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*A)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RDI
 
 | |
2IJX
 
 | |
1K6Y
 
 | | Crystal Structure of a Two-Domain Fragment of HIV-1 Integrase | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, J, Ling, H, Yang, W, Craigie, R. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein.
EMBO J., 20, 2001
|
|
2NTI
 
 | | Crystal structure of PCNA123 heterotrimer. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, BROMIDE ION, DNA polymerase sliding clamp A, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8TG1
 
 | | Caldicellulosiruptor saccharolyticus periplasmic urea-binding protein | | Descriptor: | BROMIDE ION, Extracellular ligand-binding receptor, UREA | | Authors: | Allert, M.J, Kumar, S, Wang, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure-based functional analysis reveals multiple roles and widespread use of urea-binding proteins in nitrogen metabolism
To Be Published
|
|
8FXT
 
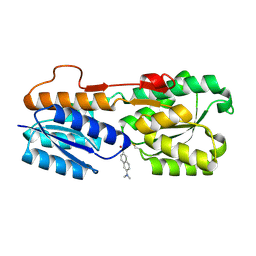 | | Escherichia coli periplasmic Glucose-Binding Protein glucose complex: Acrylodan conjugate attached at W183C | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]propan-1-one, CALCIUM ION, D-galactose/methyl-galactoside binding periplasmic protein MglB, ... | | Authors: | Allert, M.J, Kumar, S, Wang, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
8FXU
 
 | | Thermoanaerobacter thermosaccharolyticum periplasmic Glucose-Binding Protein glucose complex: Badan conjugate attached at F17C | | Descriptor: | 2-bromo-1-[6-(dimethylamino)naphthalen-2-yl]ethan-1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Allert, M.J, Kumar, S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
3E3I
 
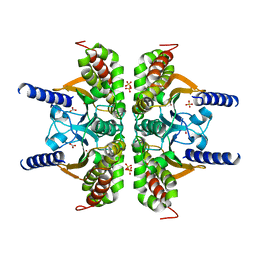 | |
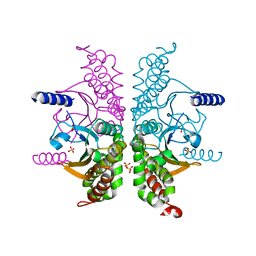
3E3G
 
 | |
3I5O
 
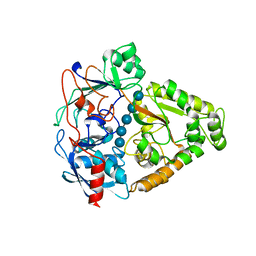 | | The X-ray crystal structure of a thermophilic cellobiose binding protein bound with cellopentaose | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cuneo, M.J, Hellinga, H.W. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Semi-specific Oligosaccharide Recognition by a Cellulose-binding Protein of Thermotoga maritima Reveals Adaptations for Functional Diversification of the Oligopeptide Periplasmic Binding Protein Fold.
J.Biol.Chem., 284, 2009
|
|
3C6Q
 
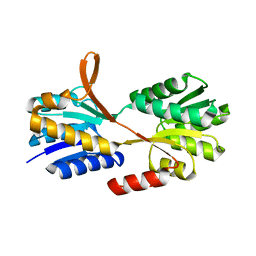 | |
3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3SV5
 
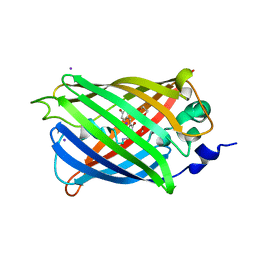 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3QEA
 
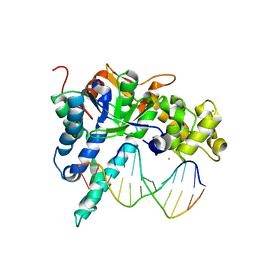 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with DNA (complex II) | | Descriptor: | BARIUM ION, DNA (5'-D(P*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QE9
 
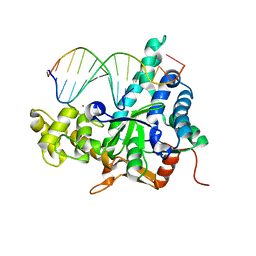 | | Crystal structure of human exonuclease 1 Exo1 (D173A) in complex with DNA (complex I) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QEB
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with DNA and Mn2+ (complex III) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3SSP
 
 | |
3SST
 
 | |
3SSV
 
 | |
3SS0
 
 | |
3SSH
 
 | |
