2N2U
 
 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | Descriptor: | OR358 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
2N75
 
 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
3DZO
 
 | | Crystal structure of a rhoptry kinase from toxoplasma gondii | | Descriptor: | MAGNESIUM ION, Rhoptry kinase domain | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Ni, S, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sibley, D, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
2HGA
 
 | | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A | | Descriptor: | Conserved protein MTH1368 | | Authors: | Liu, G, Lin, Y, Parish, D, Shen, Y, Sukumaran, D, Yee, A, Semesi, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A
TO BE PUBLISHED
|
|
2G0H
 
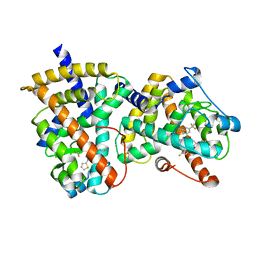 | | Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities | | Descriptor: | N-[1-(4-FLUOROPHENYL)-3-(2-THIENYL)-1H-PYRAZOL-5-YL]-3,5-BIS(TRIFLUOROMETHYL)BENZENESULFONAMIDE, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Huang, C.F, Lin, Y.T, Hsu, J.T.A, Wu, S.Y. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Drug Design of a Novel Family of PPARgamma Partial Agonists: Virtual Screening, X-ray Crystallography, and in Vitro/in Vivo Biological Activities
J.Med.Chem., 49, 2006
|
|
2G0G
 
 | | Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities | | Descriptor: | 3-FLUORO-N-[1-(4-FLUOROPHENYL)-3-(2-THIENYL)-1H-PYRAZOL-5-YL]BENZENESULFONAMIDE, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Huang, C.F, Lin, Y.T, Hsu, J.T.A, Wu, S.Y. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure-Based Drug Design of a Novel Family of PPARgamma Partial Agonists: Virtual Screening, X-ray Crystallography, and in Vitro/in Vivo Biological Activities
J.Med.Chem., 49, 2006
|
|
7F07
 
 | | Autonomous VH domain that interacts with eIF4E at the Capped mRNA Binding site. | | Descriptor: | Eukaryotic translation initiation factor 4E, VH domain (VH-DiFCAP-01) | | Authors: | Brown, C.J, Frosi, Y, Ng, S, Lin, Y.C. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
7D8B
 
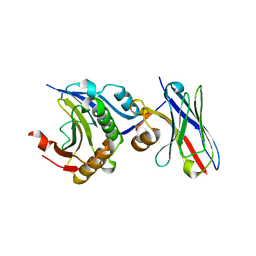 | | Engineering Disulphide-Free Autonomous Antibody VH Domains to modulate intracellular pathways | | Descriptor: | Eukaryotic translation initiation factor 4E, VH-S4 | | Authors: | Frosi, Y, Lin, Y.C, Jiang, S, Brown, C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex.
Nat Commun, 13, 2022
|
|
7D6Y
 
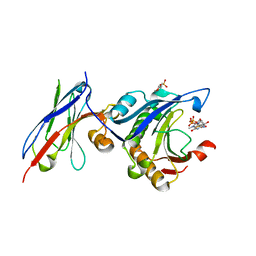 | | eIF4E in Complex with a Disulphide-Free Autonomous VH Domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Eukaryotic translation initiation factor 4E, VH Domain (1C5), ... | | Authors: | Brown, C.J, Frosi, Y, Jiang, S, Lin, Y.C. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Engineering Disulphide-Free Autonomous Antibody VH Domains to modulate intracellular pathways
To Be Published
|
|
2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | Descriptor: | Bacterial Ig-like domain, group 2 | | Authors: | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
2OEY
 
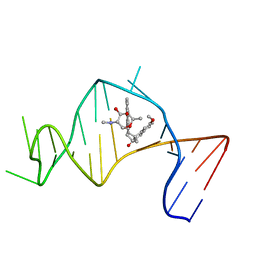 | | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA | | Descriptor: | (1R,3A'S,10'S,10A'R)-7-METHOXY-2-OXO-10',10A'-DIHYDRO-2H,3A'H-SPIRO[NAPHTHALENE-1,3'-PENTALENO[1,2-B]NAPHTHALEN]-10'-YL 2,6-DIDEOXY-2-(METHYLAMINO)-ALPHA-D-GALACTOPYRANOSIDE, DNA (25-MER) | | Authors: | Zhang, N, Lin, Y, Xiao, Z, Jones, G.B, Goldberg, I.H. | | Deposit date: | 2007-01-01 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA.
Biochemistry, 46, 2007
|
|
2O1Z
 
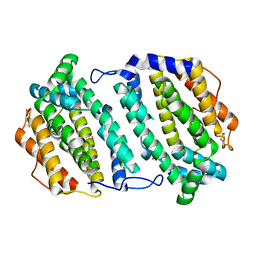 | | Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155) | | Descriptor: | FE (III) ION, Ribonucleotide Reductase Subunit R2, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Tempel, W, Qiu, W, Lew, J, Wernimont, A.K, Lin, Y.H, Hassanali, A, Melone, M, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155)
To be Published
|
|
2YP3
 
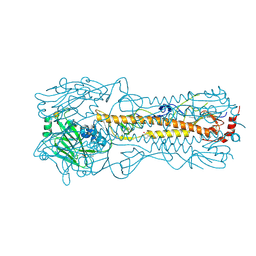 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP4
 
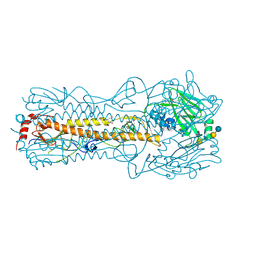 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP9
 
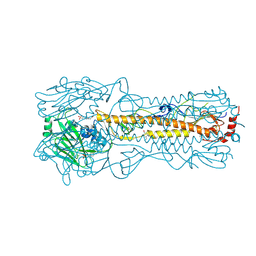 | | Haemagglutinin of 2005 Human H3N2 Virus in Complex with Avian Receptor Analogue 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP7
 
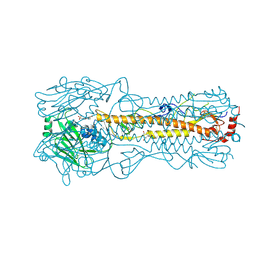 | | Haemagglutinin of 2005 Human H3N2 Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP5
 
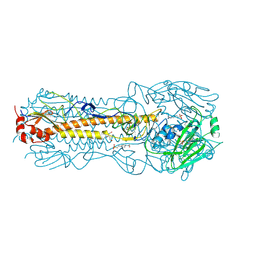 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Avian Receptor Analogue 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP2
 
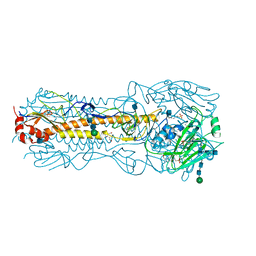 | | Haemagglutinin of 2004 Human H3N2 Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1DUJ
 
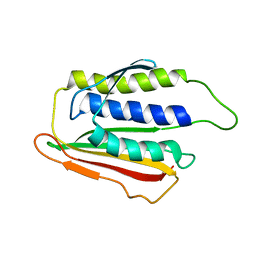 | | SOLUTION STRUCTURE OF THE SPINDLE ASSEMBLY CHECKPOINT PROTEIN HUMAN MAD2 | | Descriptor: | SPINDLE ASSEMBLY CHECKPOINT PROTEIN | | Authors: | Luo, X, Fang, G, Coldiron, M, Lin, Y, Yu, H. | | Deposit date: | 2000-01-17 | | Release date: | 2000-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20.
Nat.Struct.Biol., 7, 2000
|
|
2YP8
 
 | | Haemagglutinin of 2005 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1RHT
 
 | | 24-MER RNA HAIRPIN COAT PROTEIN BINDING SITE FOR BACTERIOPHAGE R17 (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RNA (5'-R(P*GP*GP*GP*AP*CP*UP*GP*AP*CP*GP*AP*UP*CP*AP*CP*GP*CP*AP*GP*UP*CP*UP*AP*U)-3') | | Authors: | Borer, P.N, Lin, Y, Wang, S, Roggenbuck, M.W, Gott, J.M, Uhlenbeck, O.C, Pelczer, I. | | Deposit date: | 1995-03-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Biochemistry, 34, 1995
|
|
3BYV
 
 | | Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Rhoptry kinase | | Authors: | Wernimont, A.K, Lunin, V.V, Yang, C, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Sibley, D, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
7XKX
 
 | | Crystal structure of Tpn2 | | Descriptor: | SQHop_cyclase_C domain-containing protein | | Authors: | Chang, C.Y, Stowell, E.A, Lin, Y.L, Ehrenberger, M.A, Rudolf, J.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure-guided product determination of the bacterial type II diterpene synthase Tpn2.
Commun Chem, 5, 2022
|
|
7Y62
 
 | | Crystal structure of human TFEB HLHLZ domain | | Descriptor: | Transcription factor EB | | Authors: | Yang, G, Li, P, Lin, Y, Liu, Z, Sun, H, Zhao, Z, Fang, P, Wang, J. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small-molecule drug inhibits autophagy gene expression through the central regulator TFEB.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
