3FB3
 
 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
3H80
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 83-1, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
To be Published
|
|
3F9R
 
 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
3GUE
 
 | | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Wernimont, A.K, Marino, K, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-29 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330)
To be Published
|
|
4N7R
 
 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MQ8
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-06-12 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Control over overall shape and size in de novo designed proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2HAH
 
 | | The structure of FIV 12S protease in complex with TL-3 | | Descriptor: | Protease, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Lin, Y.C, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an FIV/HIV chimeric protease complexed with the broad-based inhibitor, TL-3.
Retrovirology, 4, 2007
|
|
3EFZ
 
 | | Crystal Structure of a 14-3-3 protein from cryptosporidium parvum (cgd1_2980) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein | | Authors: | Wernimont, A.K, Dong, A, Qiu, W, Lew, J, Wasney, G.A, Vedadi, M, Kozieradzki, I, Zhao, Y, Ren, H, Alam, Z, Lin, Y.H, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
3E95
 
 | | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carrier protein, Ubiquitin-conjugating enzyme E2 | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Brokx, S, Lin, Y.H, Zhao, Y, Lew, J, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, BOuntra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Qiu, W, Brand, V.B, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a
TO BE PUBLISHED
|
|
3I3G
 
 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
3E5R
 
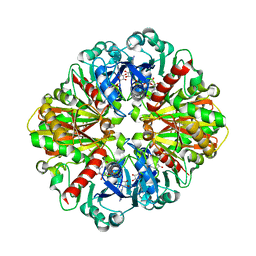 | |
3E6A
 
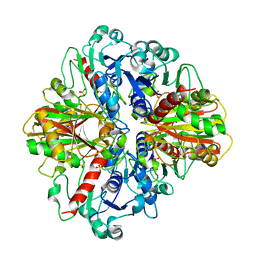 | |
3EB0
 
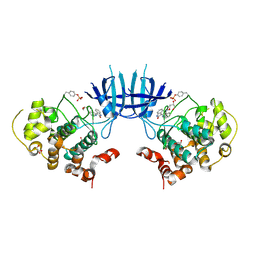 | | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, GLYCEROL, Putative uncharacterized protein | | Authors: | Wernimont, A.K, Fedorov, O, Lam, A, Ali, A, Zhao, Y, Lew, J, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804
TO BE PUBLISHED
|
|
2D9S
 
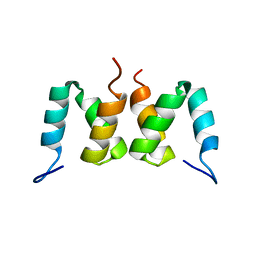 | | Solution structure of RSGI RUH-049, a UBA domain from mouse cDNA | | Descriptor: | CBL E3 ubiquitin protein ligase | | Authors: | Hamada, T, Hirota, H, Lin, Y.-J, Guntert, P, Kurosaki, C, Izumi, K, Yoshida, M, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-049, a UBA domain from mouse cDNA
To be Published
|
|
7CEN
 
 | |
7CEZ
 
 | |
7WX6
 
 | | A Legionella acetyltransferase VipF | | Descriptor: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3SHB
 
 | | Crystal Structure of PHD Domain of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 peptide, ZINC ION | | Authors: | Hu, L, Li, Z, Wang, P, Lin, Y, Xu, Y. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2.
Cell Res., 2011
|
|
7XQL
 
 | | complex structure of LegA15 with GNP | | Descriptor: | Ankyrin repeat-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
2N2T
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303 | | Descriptor: | OR303 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303
To be Published
|
|
2N3Z
 
 | | Solution NMR Structure of de novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | OR446 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-15 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
8IMZ
 
 | | Cryo-EM structure of mouse Piezo1-MDFIC complex (composite map) | | Descriptor: | MyoD family inhibitor domain-containing protein, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhou, Z, Ma, X, Lin, Y, Cheng, D, Bavi, N, Li, J.V, Sutton, D, Yao, M, Harvey, N, Corry, B, Zhang, Y, Cox, C.D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels.
Science, 381, 2023
|
|
7WX7
 
 | | complex of a legionella acetyltransferase VipF and COA/ACO | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX5
 
 | | a Legionella acetyltransferase effector VipF | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3HJC
 
 | | Crystal structure of the carboxy-terminal domain of HSP90 from Leishmania major, LmjF33.0312 | | Descriptor: | Heat shock protein 83-1, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Walker, J, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the middle and carboxy-terminal domain of HSP90 from Leishmania major, LMJF33.0312
To be Published
|
|
